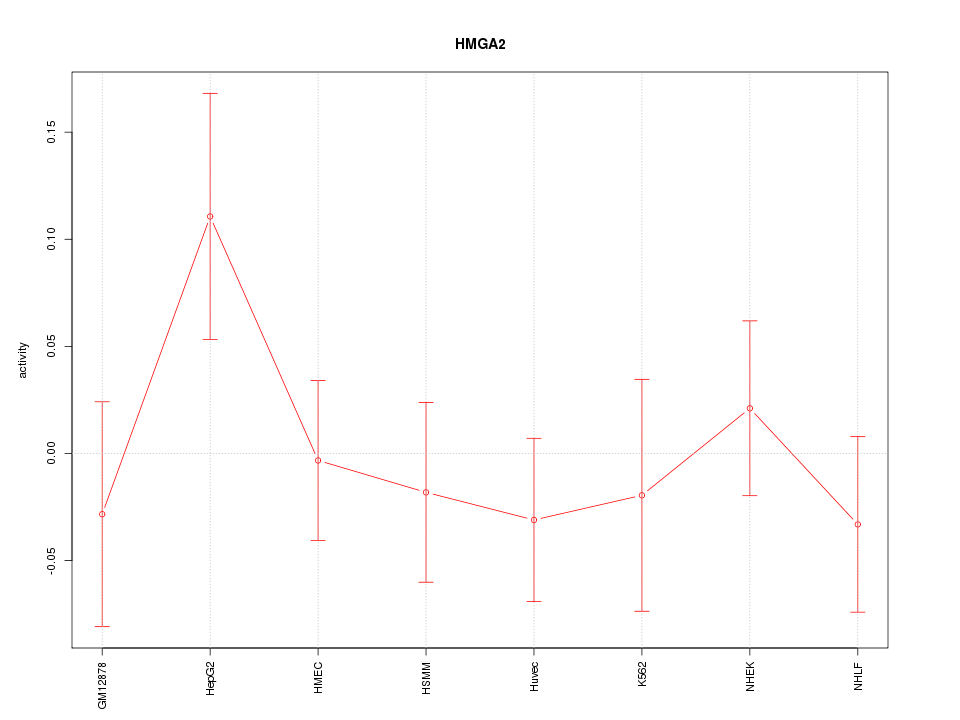
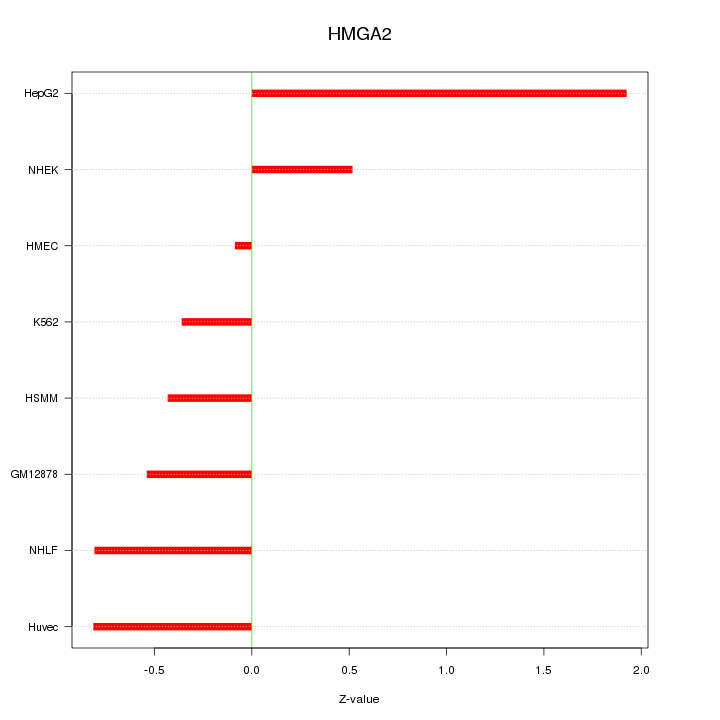
Motif ID: HMGA2
Z-value: 0.859
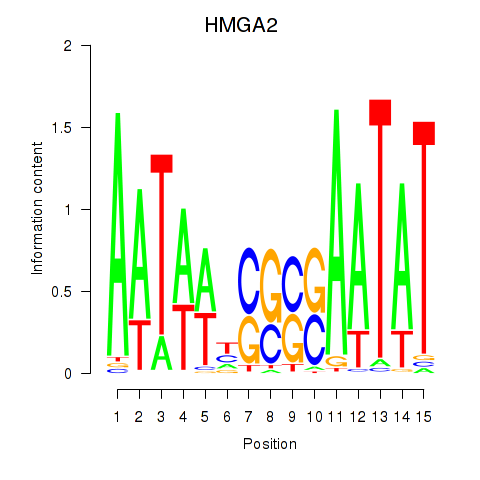
Transcription factors associated with HMGA2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HMGA2 | ENSG00000149948.9 | HMGA2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.4 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.3 | 1.0 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.3 | 3.0 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.3 | 2.4 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 | 0.7 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.2 | 1.2 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.2 | 0.7 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.1 | 1.5 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 2.4 | GO:0006693 | prostaglandin metabolic process(GO:0006693) |
| 0.1 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:0048070 | regulation of developmental pigmentation(GO:0048070) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0047026 | androsterone dehydrogenase (A-specific) activity(GO:0047026) |
| 0.3 | 1.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.3 | 1.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.2 | 2.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 3.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 1.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 3.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |