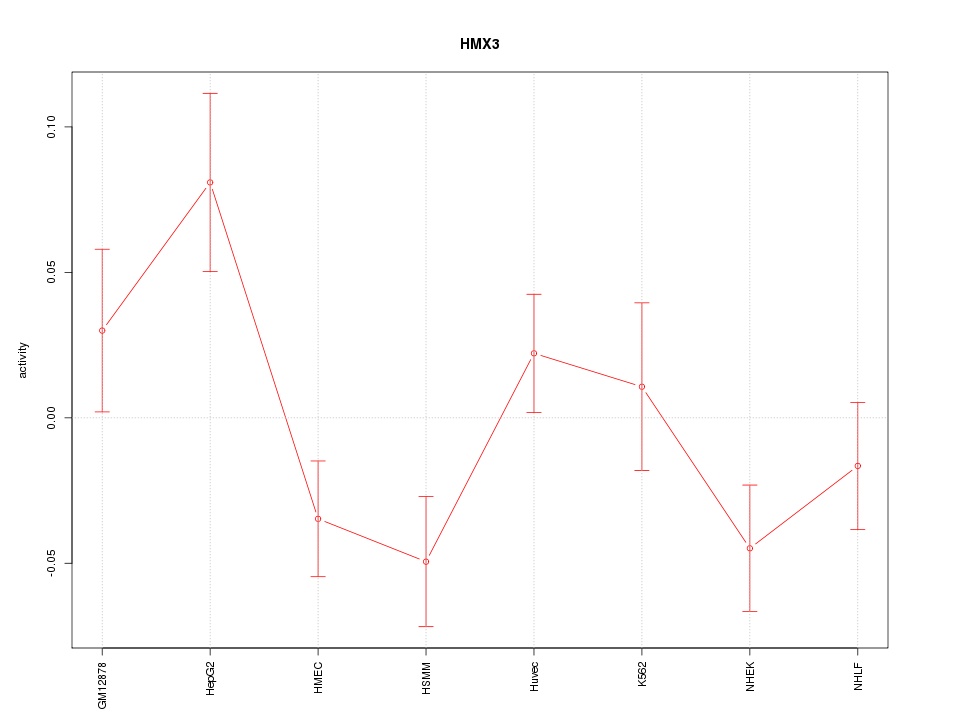
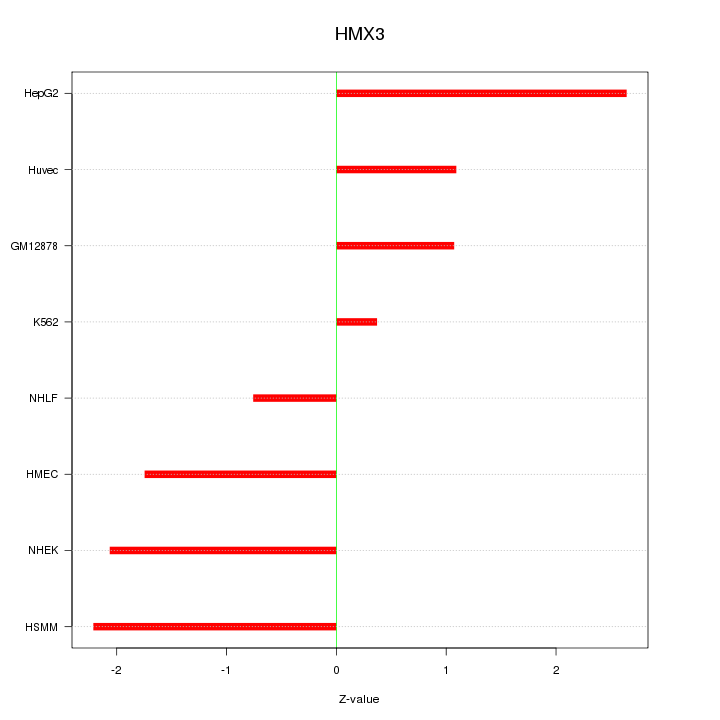
Motif ID: HMX3
Z-value: 1.667
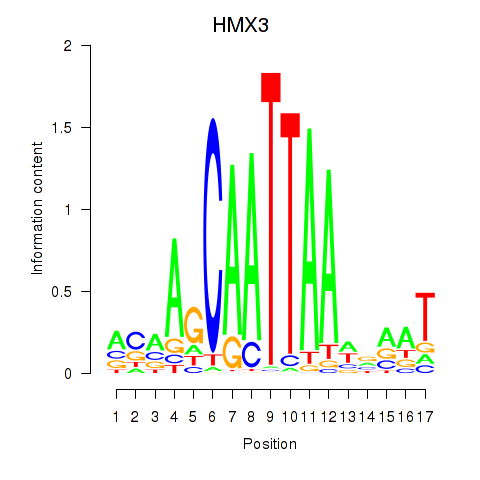
Transcription factors associated with HMX3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HMX3 | ENSG00000188620.9 | HMX3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.7 | 4.1 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.6 | 5.2 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.5 | 1.6 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.5 | 1.6 | GO:1903961 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.5 | 1.4 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.3 | 0.8 | GO:0014063 | regulation of serotonin secretion(GO:0014062) negative regulation of serotonin secretion(GO:0014063) |
| 0.2 | 2.2 | GO:0034382 | chylomicron remnant clearance(GO:0034382) phosphatidylcholine catabolic process(GO:0034638) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.2 | 0.7 | GO:0002431 | Fc receptor mediated stimulatory signaling pathway(GO:0002431) regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.2 | 1.1 | GO:0042905 | retinal metabolic process(GO:0042574) 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 1.4 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 0.2 | GO:0070445 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.2 | 0.6 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.2 | 0.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 2.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 0.7 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.2 | 0.7 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.2 | 0.7 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.2 | 1.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 0.6 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.2 | 0.5 | GO:0034126 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) positive regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034126) |
| 0.1 | 1.5 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 3.2 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.5 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 0.6 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.4 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 1.7 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.4 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.3 | GO:0071639 | positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.1 | 0.5 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.7 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.1 | 0.7 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.4 | GO:0033622 | integrin activation(GO:0033622) regulation of integrin activation(GO:0033623) positive regulation of integrin activation(GO:0033625) |
| 0.1 | 0.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.3 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.1 | 0.2 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 0.6 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 1.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.8 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 1.9 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 0.7 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.1 | 0.6 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) |
| 0.1 | 0.2 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.2 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.1 | 0.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.4 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 0.4 | GO:0043316 | natural killer cell activation involved in immune response(GO:0002323) cytotoxic T cell degranulation(GO:0043316) natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.5 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 | 0.2 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.1 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 1.1 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.1 | 3.3 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.1 | 0.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.3 | GO:0060744 | thelarche(GO:0042695) development of secondary female sexual characteristics(GO:0046543) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.2 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.4 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.8 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.3 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.5 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 2.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.6 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.2 | GO:1902117 | positive regulation of centrosome cycle(GO:0046607) positive regulation of organelle assembly(GO:1902117) |
| 0.0 | 0.4 | GO:0030147 | obsolete natriuresis(GO:0030147) |
| 0.0 | 0.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.3 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.3 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.6 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.9 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 | 0.4 | GO:0032464 | regulation of protein homooligomerization(GO:0032462) positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 1.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.4 | GO:1902622 | regulation of neutrophil chemotaxis(GO:0090022) regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.1 | GO:0048865 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.7 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.2 | GO:0099515 | nuclear migration along microfilament(GO:0031022) actin filament-based transport(GO:0099515) |
| 0.0 | 0.4 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.1 | GO:0010430 | ethanol catabolic process(GO:0006068) fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.5 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.2 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) |
| 0.0 | 0.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.3 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.7 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.1 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.0 | 0.4 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.6 | GO:0045909 | positive regulation of vasodilation(GO:0045909) |
| 0.0 | 1.0 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 0.5 | GO:0030260 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.4 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.8 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.2 | GO:0051294 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.4 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.1 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.2 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.3 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.7 | GO:0009067 | aspartate family amino acid biosynthetic process(GO:0009067) |
| 0.0 | 0.2 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.4 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.3 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.7 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.6 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0034101 | erythrocyte differentiation(GO:0030218) erythrocyte homeostasis(GO:0034101) |
| 0.0 | 1.7 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 0.1 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.6 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.3 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.2 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.1 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.6 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.0 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0034372 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.7 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0015893 | drug transport(GO:0015893) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.3 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.1 | GO:0001916 | positive regulation of T cell mediated cytotoxicity(GO:0001916) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.7 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 3.6 | GO:0007067 | mitotic nuclear division(GO:0007067) |
| 0.0 | 0.2 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.1 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.4 | GO:0050663 | cytokine secretion(GO:0050663) |
| 0.0 | 0.8 | GO:0071466 | xenobiotic metabolic process(GO:0006805) response to xenobiotic stimulus(GO:0009410) cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.2 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.2 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 0.1 | GO:0047496 | vesicle transport along microtubule(GO:0047496) vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.4 | GO:0051781 | positive regulation of cell division(GO:0051781) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 6.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 1.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.8 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.3 | GO:0033011 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.1 | 1.1 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.1 | 0.5 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 1.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 2.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.8 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 2.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.3 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.7 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.4 | GO:0097525 | spliceosomal snRNP complex(GO:0097525) |
| 0.0 | 0.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 2.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 6.3 | GO:0005792 | obsolete microsome(GO:0005792) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.7 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.7 | GO:0030141 | secretory granule(GO:0030141) secretory vesicle(GO:0099503) |
| 0.0 | 1.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 4.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.9 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.5 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.2 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.6 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.4 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 1.0 | 4.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.3 | 2.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 1.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 1.2 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.3 | 10.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 0.8 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.2 | 0.7 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) |
| 0.2 | 0.7 | GO:0015247 | aminophospholipid transporter activity(GO:0015247) |
| 0.2 | 1.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 0.6 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.2 | 3.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.2 | 1.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 2.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 1.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.2 | 0.7 | GO:0008907 | integrase activity(GO:0008907) |
| 0.2 | 0.5 | GO:0047042 | androsterone dehydrogenase (B-specific) activity(GO:0047042) |
| 0.2 | 2.7 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.2 | 0.5 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.2 | 0.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 1.5 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 1.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 1.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.7 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 1.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 1.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.6 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 2.3 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.1 | 0.9 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 0.7 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.1 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.2 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 1.6 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) fatty acid ligase activity(GO:0015645) |
| 0.1 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.4 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.2 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.1 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.2 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 0.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) SAM domain binding(GO:0032093) |
| 0.1 | 0.2 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 2.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 2.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 1.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.7 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.4 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.2 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 2.5 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.3 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.5 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.4 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.4 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.1 | GO:0046980 | oligopeptide-transporting ATPase activity(GO:0015421) tapasin binding(GO:0046980) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 1.3 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.0 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.7 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.1 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.4 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.8 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.3 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |