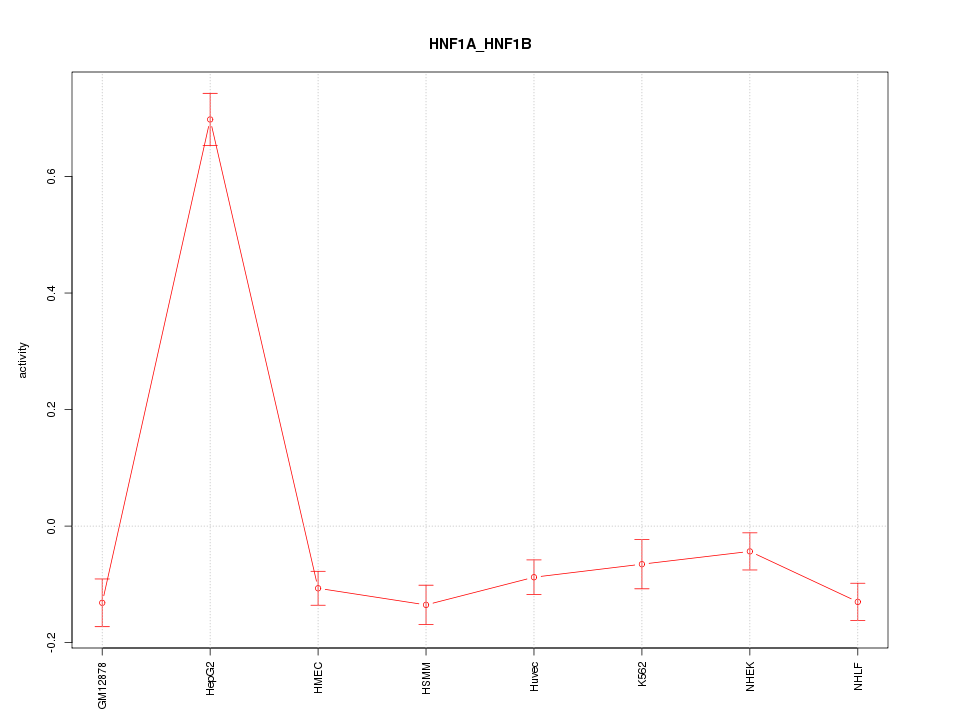
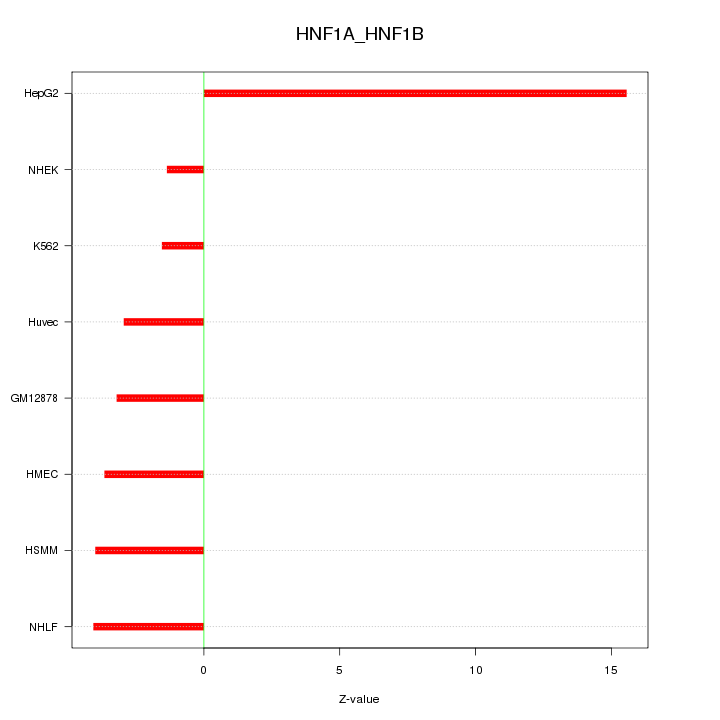
Motif ID: HNF1A_HNF1B
Z-value: 6.240
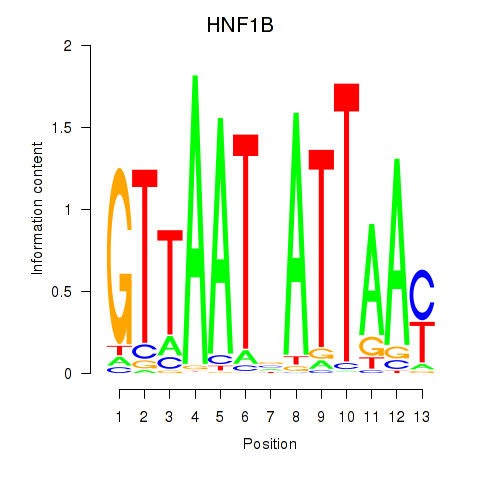
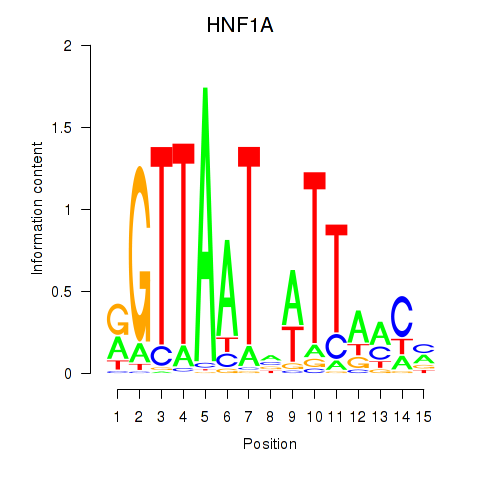
Transcription factors associated with HNF1A_HNF1B:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HNF1A | ENSG00000135100.13 | HNF1A |
| HNF1B | ENSG00000108753.8 | HNF1B |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 66.8 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 6.4 | 25.5 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 6.1 | 60.7 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 5.5 | 16.5 | GO:0001907 | cytolysis by symbiont of host cells(GO:0001897) killing by symbiont of host cells(GO:0001907) hemolysis by symbiont of host erythrocytes(GO:0019836) disruption by symbiont of host cell(GO:0044004) hemolysis in other organism(GO:0044179) positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) cytolysis in other organism(GO:0051715) cytolysis in other organism involved in symbiotic interaction(GO:0051801) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 4.7 | 18.8 | GO:0033986 | response to methanol(GO:0033986) response to chromate(GO:0046687) |
| 4.1 | 12.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 4.0 | 40.0 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 3.4 | 27.1 | GO:0034695 | response to prostaglandin E(GO:0034695) |
| 3.2 | 12.7 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 2.8 | 16.5 | GO:0006562 | proline catabolic process(GO:0006562) |
| 2.4 | 9.6 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 2.3 | 11.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 2.0 | 113.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 1.9 | 5.8 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 1.8 | 8.9 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 1.5 | 13.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 1.3 | 5.1 | GO:0033622 | integrin activation(GO:0033622) regulation of integrin activation(GO:0033623) positive regulation of integrin activation(GO:0033625) |
| 1.3 | 12.8 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 1.3 | 8.8 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 1.1 | 5.4 | GO:1903301 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 1.0 | 3.1 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 1.0 | 3.9 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.8 | 3.3 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.7 | 5.6 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.6 | 29.9 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.5 | 17.4 | GO:0006693 | prostaglandin metabolic process(GO:0006693) |
| 0.5 | 2.2 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.5 | 51.8 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.5 | 2.4 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.4 | 1.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.4 | 2.8 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.4 | 5.3 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.4 | 2.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.3 | 4.9 | GO:0017158 | regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.3 | 17.1 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.3 | 5.9 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 28.5 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.2 | 0.7 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.2 | 14.9 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 5.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 6.3 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.1 | 9.0 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 3.3 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 9.2 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.1 | 0.7 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 10.5 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 2.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.0 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.8 | GO:0051925 | obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 | 3.3 | GO:0006402 | mRNA catabolic process(GO:0006402) |
| 0.0 | 7.3 | GO:0015711 | organic anion transport(GO:0015711) |
| 0.0 | 1.6 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.8 | GO:0001707 | mesoderm formation(GO:0001707) |
| 0.0 | 2.9 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 2.0 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 0.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.8 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 1.3 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 64.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 4.1 | 12.4 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 3.0 | 27.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 2.2 | 18.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 2.1 | 12.8 | GO:0045179 | apical cortex(GO:0045179) |
| 1.9 | 41.9 | GO:0042627 | chylomicron(GO:0042627) |
| 1.9 | 11.4 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.9 | 6.3 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.8 | 3.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.8 | 9.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.6 | 14.9 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.6 | 15.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.5 | 35.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.4 | 3.9 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.3 | 1.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 1.2 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.3 | 7.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 3.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 1.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 163.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 5.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 2.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 11.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 6.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 3.1 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 5.1 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 68.1 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 1.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 3.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 14.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 16.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 4.9 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 1.7 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 23.7 | GO:0005624 | obsolete membrane fraction(GO:0005624) |
| 0.0 | 10.8 | GO:0005625 | obsolete soluble fraction(GO:0005625) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 6.1 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 3.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 19.9 | GO:0005654 | nucleoplasm(GO:0005654) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 41.9 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 5.8 | 17.4 | GO:0047718 | indanol dehydrogenase activity(GO:0047718) |
| 4.1 | 16.5 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 3.7 | 14.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 3.2 | 12.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 2.6 | 25.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 2.3 | 49.8 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 2.0 | 18.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 1.7 | 6.9 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.5 | 16.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 1.4 | 11.4 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 1.4 | 9.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 1.3 | 16.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 1.3 | 64.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 1.3 | 19.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 1.1 | 31.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 1.1 | 12.4 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 1.0 | 5.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.9 | 11.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.9 | 93.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.8 | 7.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.8 | 3.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.7 | 5.4 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.7 | 12.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.7 | 5.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.6 | 2.2 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.5 | 24.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.4 | 4.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 12.4 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.3 | 5.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 5.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 1.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.2 | 3.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 0.7 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 3.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.2 | 0.8 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) glycine:sodium symporter activity(GO:0015375) |
| 0.2 | 8.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 4.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 12.8 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 5.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 9.7 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.1 | 1.2 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.1 | 0.7 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 8.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 3.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 8.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 2.8 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 6.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 2.4 | GO:0010843 | obsolete promoter binding(GO:0010843) |
| 0.0 | 0.2 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 31.7 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
| 0.5 | 11.0 | ST_INTERLEUKIN_4_PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.2 | 7.9 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.2 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |