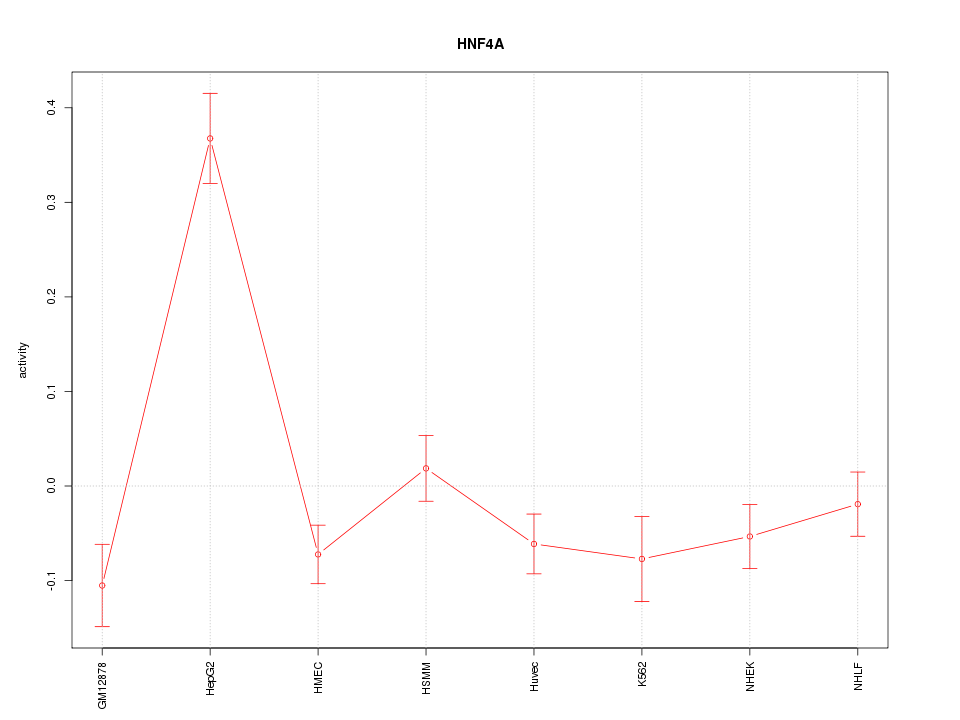
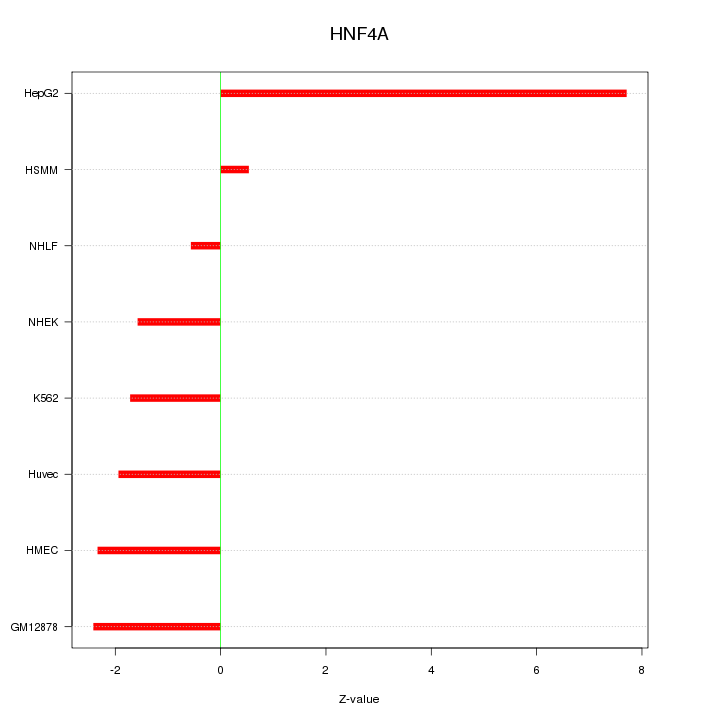
Motif ID: HNF4A
Z-value: 3.173
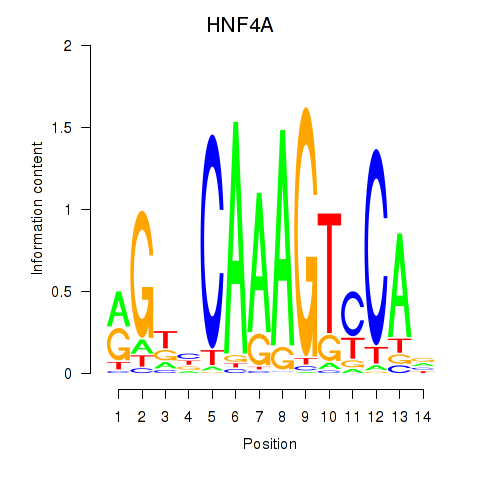
Transcription factors associated with HNF4A:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HNF4A | ENSG00000101076.12 | HNF4A |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 25.2 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) positive regulation of phospholipid catabolic process(GO:0060697) |
| 2.9 | 8.8 | GO:2000910 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 2.4 | 9.7 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 1.7 | 10.4 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 1.1 | 7.9 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 1.0 | 7.9 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 1.0 | 4.8 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.9 | 5.2 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.8 | 2.4 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.7 | 3.4 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.7 | 6.8 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.6 | 2.4 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.6 | 2.9 | GO:0035444 | nickel cation transport(GO:0015675) vanadium ion transport(GO:0015676) ferrous iron transport(GO:0015684) lead ion transport(GO:0015692) nickel cation transmembrane transport(GO:0035444) ferrous iron import(GO:0070627) iron ion import(GO:0097286) |
| 0.5 | 6.8 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.4 | 4.0 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.4 | 2.3 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.4 | 5.9 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.4 | 20.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.3 | 1.0 | GO:0002575 | basophil chemotaxis(GO:0002575) |
| 0.3 | 1.3 | GO:0046778 | modification by virus of host mRNA processing(GO:0046778) |
| 0.3 | 5.6 | GO:0015838 | amino-acid betaine transport(GO:0015838) carnitine transport(GO:0015879) drug transport(GO:0015893) |
| 0.3 | 2.5 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.3 | 0.8 | GO:0060061 | regulation of collateral sprouting(GO:0048670) Spemann organizer formation(GO:0060061) |
| 0.3 | 14.2 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.2 | 1.2 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.2 | 18.7 | GO:0031668 | cellular response to extracellular stimulus(GO:0031668) |
| 0.2 | 0.8 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 2.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.2 | 1.0 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 4.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.1 | 2.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 1.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.9 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 0.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 2.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 2.6 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.1 | 5.8 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 1.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 0.6 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 0.2 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.2 | GO:0043276 | anoikis(GO:0043276) |
| 0.1 | 0.8 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 3.6 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 12.4 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 4.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.3 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 3.2 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 6.1 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) |
| 0.0 | 0.1 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.0 | 7.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.0 | 0.2 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.4 | GO:0072332 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.2 | GO:0006706 | steroid catabolic process(GO:0006706) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 34.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.3 | 5.2 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 1.0 | 5.9 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.7 | 8.8 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.7 | 7.9 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.6 | 2.9 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 11.0 | GO:0005903 | brush border(GO:0005903) |
| 0.2 | 0.8 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.2 | 5.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 2.4 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 4.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 5.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 4.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 7.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 4.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 32.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 5.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 3.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.8 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 18.5 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 6.1 | GO:0005625 | obsolete soluble fraction(GO:0005625) |
| 0.0 | 20.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.3 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 25.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 2.3 | 6.8 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 2.2 | 8.8 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) high-density lipoprotein particle receptor binding(GO:0070653) |
| 2.1 | 18.7 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 1.5 | 9.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 1.3 | 5.2 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 1.2 | 4.8 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.1 | 3.4 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.8 | 9.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.8 | 19.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.6 | 2.9 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) ferrous iron transmembrane transporter activity(GO:0015093) lead ion transmembrane transporter activity(GO:0015094) nickel cation transmembrane transporter activity(GO:0015099) vanadium ion transmembrane transporter activity(GO:0015100) chromium ion transmembrane transporter activity(GO:0070835) |
| 0.6 | 2.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.5 | 14.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.3 | 5.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 1.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.2 | 1.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 2.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.2 | 2.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 2.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.1 | 0.8 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.1 | 0.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 19.9 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 2.4 | GO:0001228 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.6 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.8 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 5.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.6 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.1 | 0.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 5.8 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 2.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 4.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 6.5 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 7.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0030249 | cyclase regulator activity(GO:0010851) guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 9.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 2.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.3 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 5.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.6 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.2 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 1.3 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.2 | GO:0030275 | LRR domain binding(GO:0030275) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 15.4 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
| 0.0 | 1.0 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |