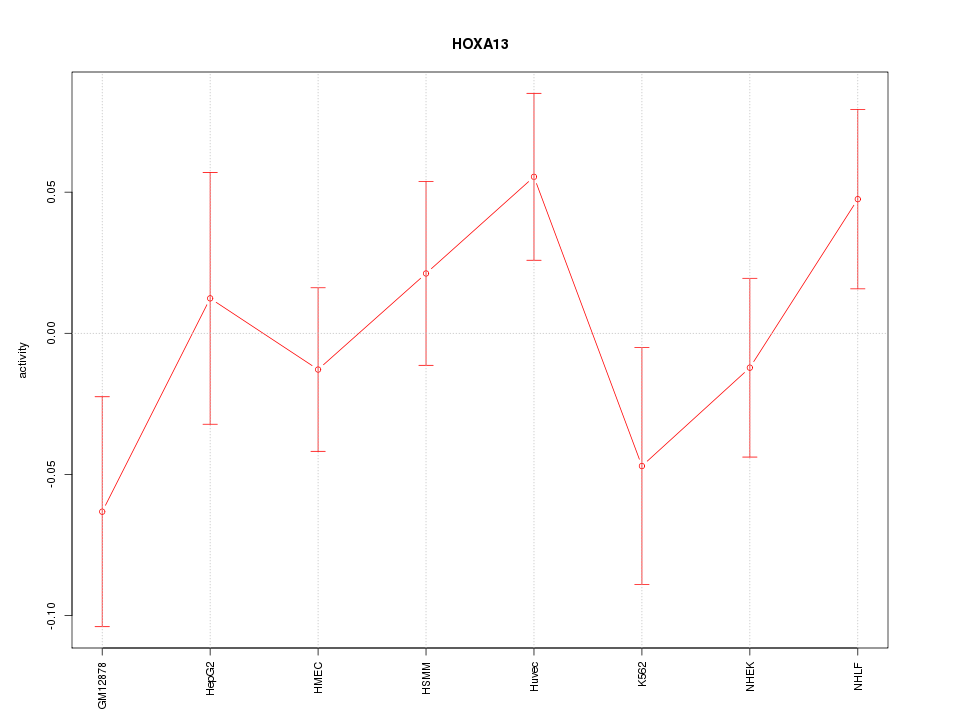
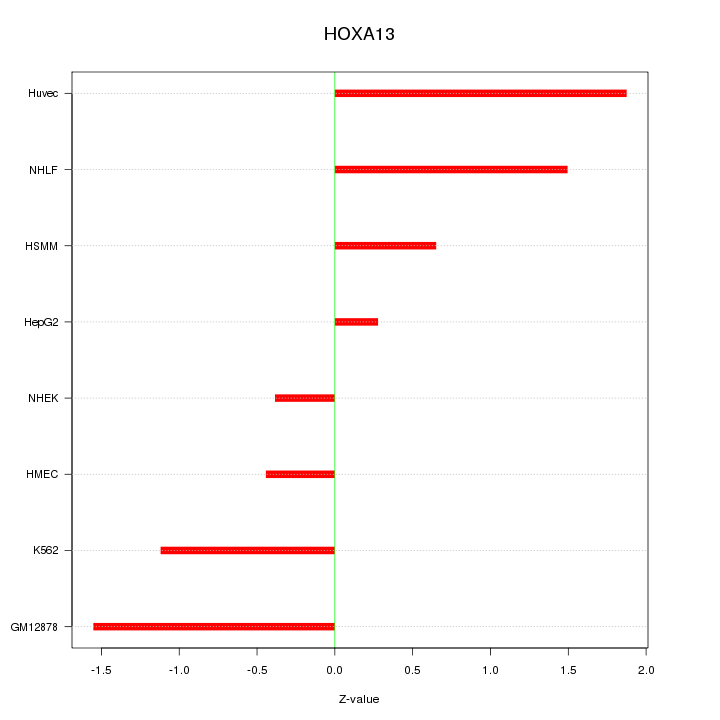
Motif ID: HOXA13
Z-value: 1.132
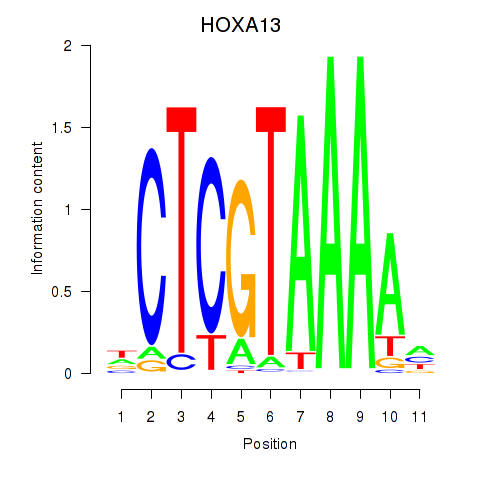
Transcription factors associated with HOXA13:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HOXA13 | ENSG00000106031.6 | HOXA13 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0051503 | purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) |
| 0.6 | 2.3 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.5 | 3.7 | GO:0060414 | aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 | 1.2 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.4 | 3.5 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.4 | 1.1 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.3 | 3.1 | GO:0097435 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.2 | 0.7 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.2 | 2.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 1.5 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.2 | 0.6 | GO:0001907 | cytolysis by symbiont of host cells(GO:0001897) killing by symbiont of host cells(GO:0001907) hemolysis by symbiont of host erythrocytes(GO:0019836) disruption by symbiont of host cell(GO:0044004) hemolysis in other organism(GO:0044179) positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) cytolysis in other organism(GO:0051715) cytolysis in other organism involved in symbiotic interaction(GO:0051801) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.2 | 1.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.2 | 0.7 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.1 | 0.6 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.4 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.4 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.3 | GO:0045578 | progesterone secretion(GO:0042701) negative regulation of B cell differentiation(GO:0045578) regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.8 | GO:0060100 | regulation of phagocytosis, engulfment(GO:0060099) positive regulation of phagocytosis, engulfment(GO:0060100) regulation of membrane invagination(GO:1905153) positive regulation of membrane invagination(GO:1905155) |
| 0.1 | 2.1 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.1 | 1.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.3 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 1.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.2 | GO:0070942 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) |
| 0.0 | 0.6 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.4 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 1.0 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.9 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 1.0 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) |
| 0.0 | 0.2 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 1.5 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.8 | GO:0042094 | interleukin-2 biosynthetic process(GO:0042094) |
| 0.0 | 1.7 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.1 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.3 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.0 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.1 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.4 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.3 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.0 | 1.7 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 3.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 2.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 2.7 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 0.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 3.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.6 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.3 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 1.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 3.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.7 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 4.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0032420 | stereocilium(GO:0032420) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.5 | 1.6 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.3 | 3.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 2.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 1.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 1.0 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.6 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.3 | GO:0048184 | obsolete follistatin binding(GO:0048184) |
| 0.1 | 3.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 2.2 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 0.3 | GO:0004608 | phosphatidylethanolamine N-methyltransferase activity(GO:0004608) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.4 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.7 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.1 | 2.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.1 | 0.4 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 1.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.8 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 3.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0004741 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 1.5 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 2.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.2 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |