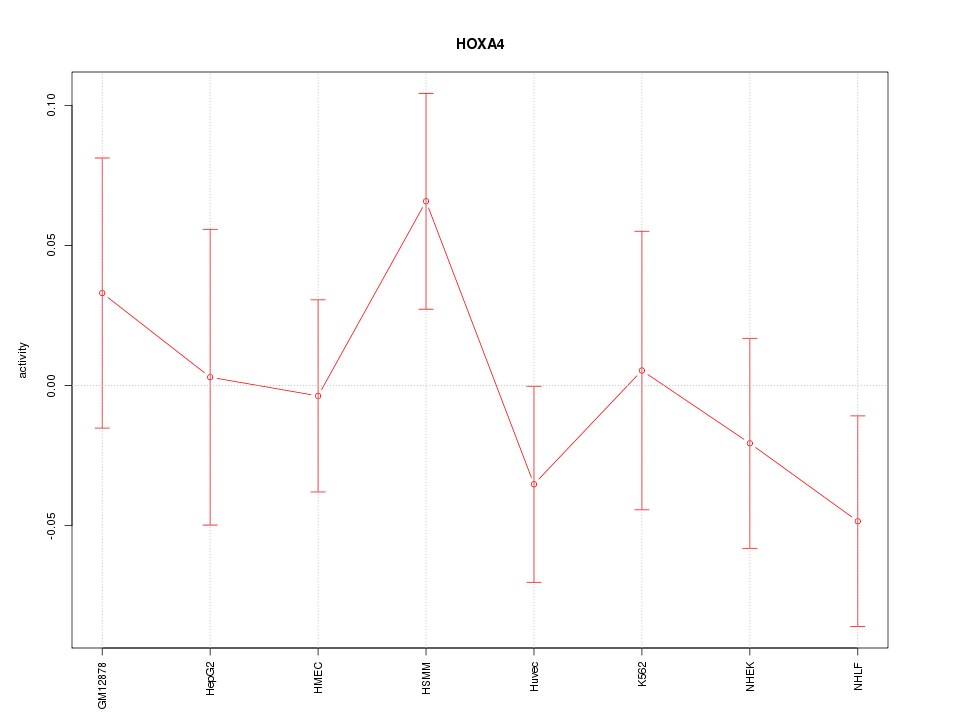
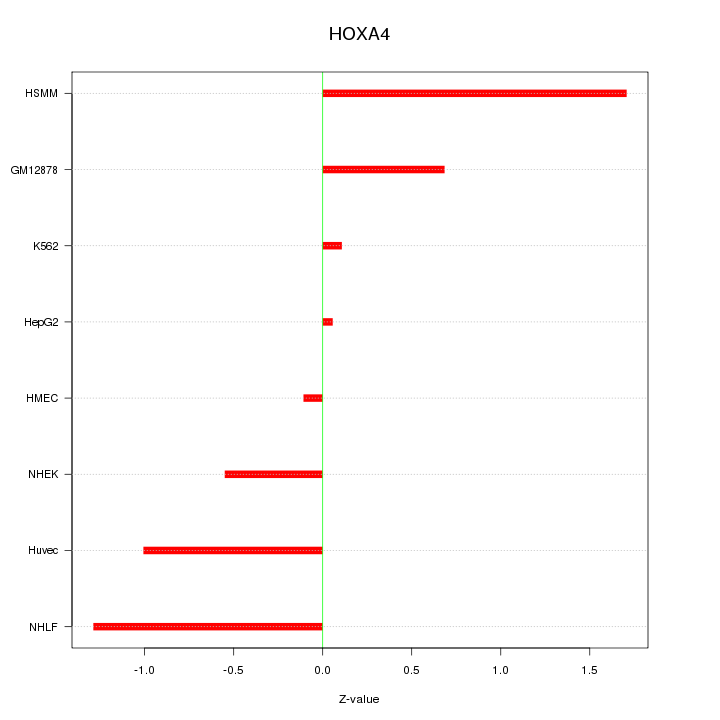
Motif ID: HOXA4
Z-value: 0.893
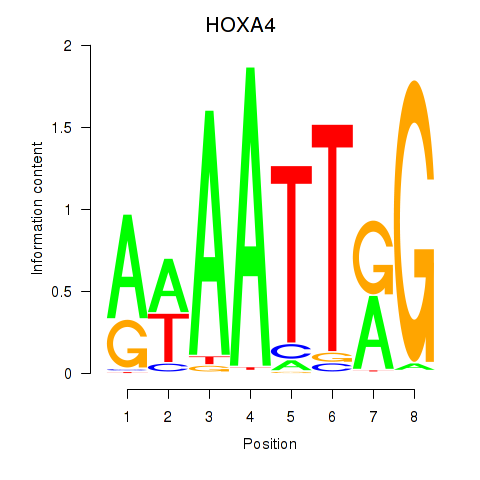
Transcription factors associated with HOXA4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HOXA4 | ENSG00000197576.9 | HOXA4 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:1903115 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.4 | 1.1 | GO:0014889 | muscle atrophy(GO:0014889) regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.3 | 1.2 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.1 | 0.3 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 | 0.2 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.3 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) menarche(GO:0042696) |
| 0.0 | 0.1 | GO:2000644 | low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) regulation of receptor catabolic process(GO:2000644) |
| 0.0 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.6 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 1.8 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.8 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 0.6 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.0 | 0.4 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.3 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.2 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:1903301 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 2.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 1.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.0 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0001228 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |