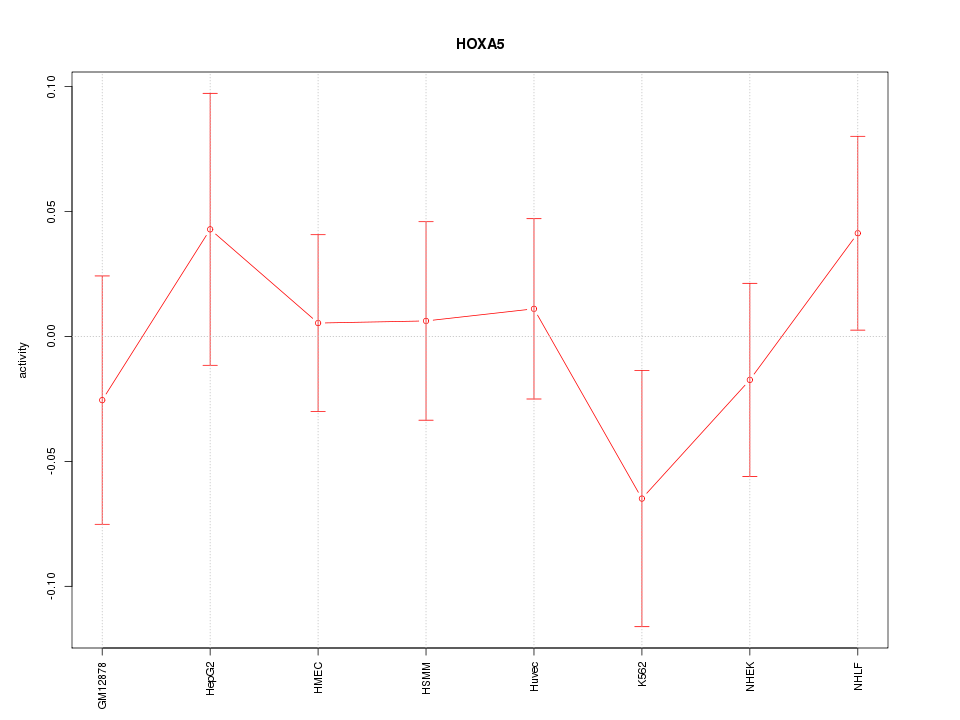
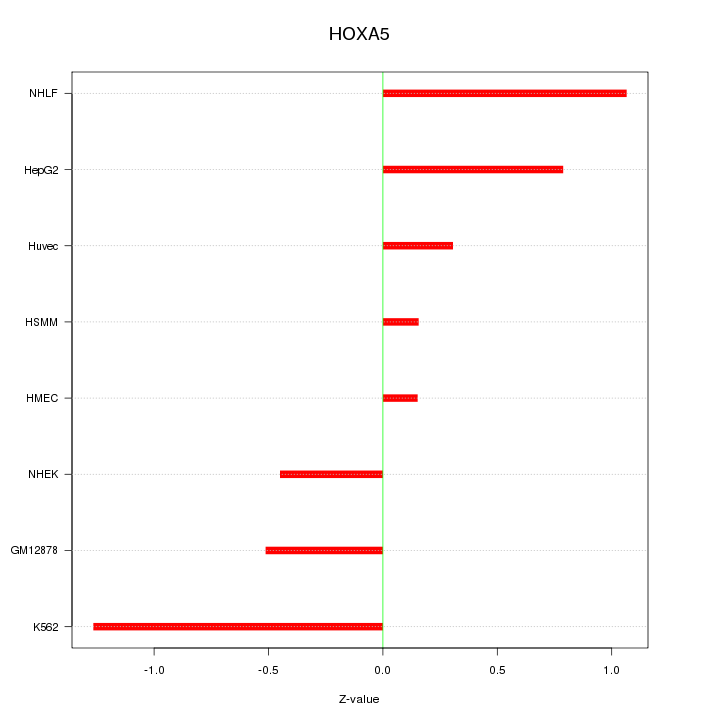
Motif ID: HOXA5
Z-value: 0.704
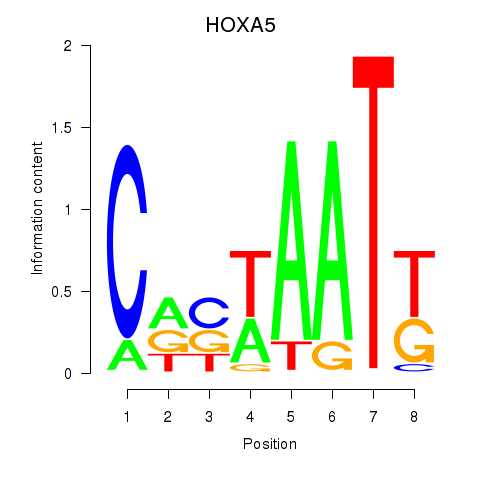
Transcription factors associated with HOXA5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HOXA5 | ENSG00000106004.4 | HOXA5 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.5 | 1.6 | GO:0001907 | cytolysis by symbiont of host cells(GO:0001897) killing by symbiont of host cells(GO:0001907) hemolysis by symbiont of host erythrocytes(GO:0019836) disruption by symbiont of host cell(GO:0044004) hemolysis in other organism(GO:0044179) positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) cytolysis in other organism(GO:0051715) cytolysis in other organism involved in symbiotic interaction(GO:0051801) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.4 | 1.3 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.4 | 0.8 | GO:0071504 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.3 | 0.9 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.2 | 0.8 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 1.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.9 | GO:0046618 | drug export(GO:0046618) |
| 0.1 | 0.5 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) long-term synaptic potentiation(GO:0060291) |
| 0.1 | 0.6 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) regulation of cholesterol transporter activity(GO:0060694) |
| 0.1 | 1.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.3 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.1 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.1 | 0.6 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.2 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.0 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0071801 | podosome assembly(GO:0071800) regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 1.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.7 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.5 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.4 | 1.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 1.0 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 1.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.5 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 1.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 1.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.9 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.1 | 0.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.0 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.5 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.3 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 0.6 | GO:1901567 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.1 | 1.9 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 1.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |