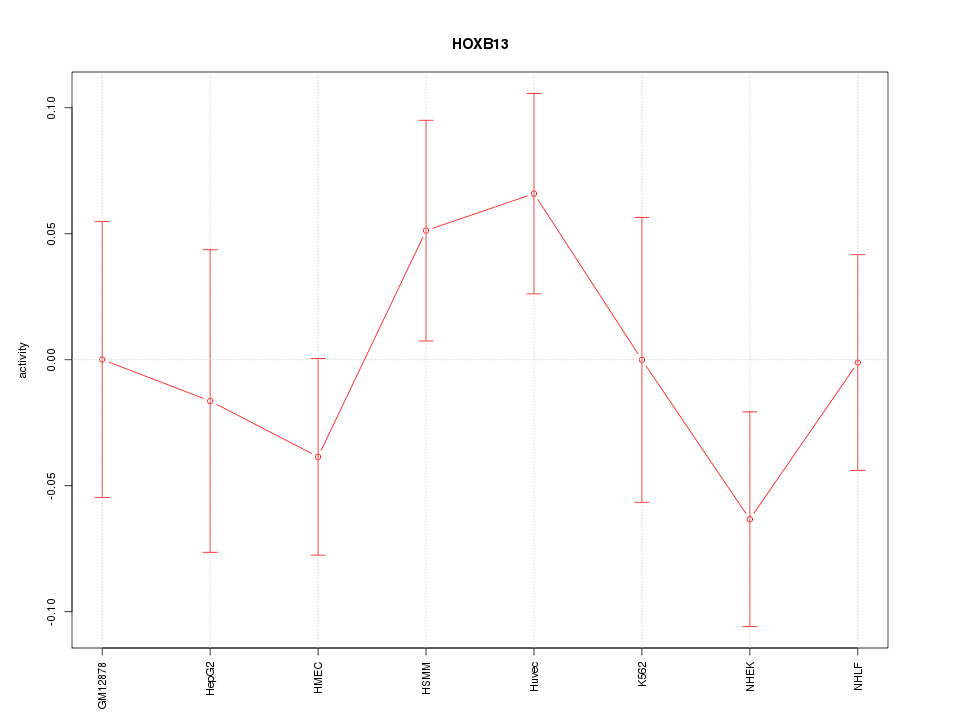
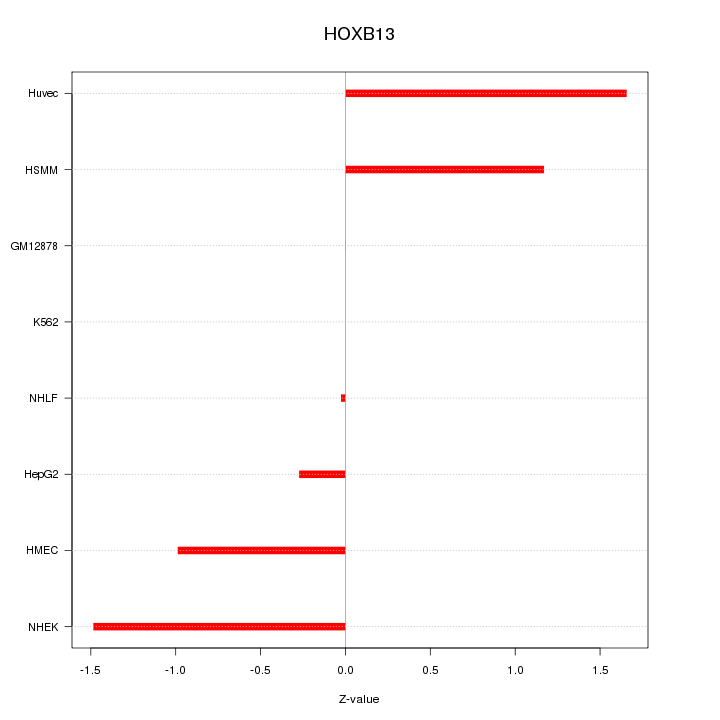
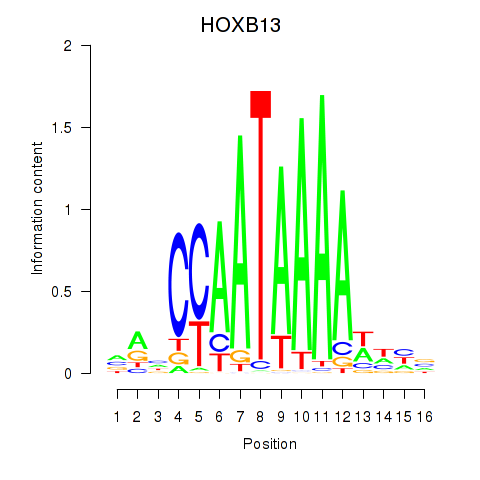
Motif ID: HOXB13
Z-value: 0.960
Transcription factors associated with HOXB13:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HOXB13 | ENSG00000159184.7 | HOXB13 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.2 | 0.6 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.1 | 0.4 | GO:0048170 | optic nerve morphogenesis(GO:0021631) positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.4 | GO:0060394 | obsolete positive regulation of S phase of mitotic cell cycle(GO:0045750) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.4 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.2 | GO:0060969 | negative regulation of gene silencing(GO:0060969) |
| 0.0 | 3.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 1.1 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.8 | GO:0017156 | calcium ion regulated exocytosis(GO:0017156) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 1.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.6 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.1 | 1.6 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 1.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) axon guidance receptor activity(GO:0008046) |
| 0.0 | 1.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.4 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 1.5 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |