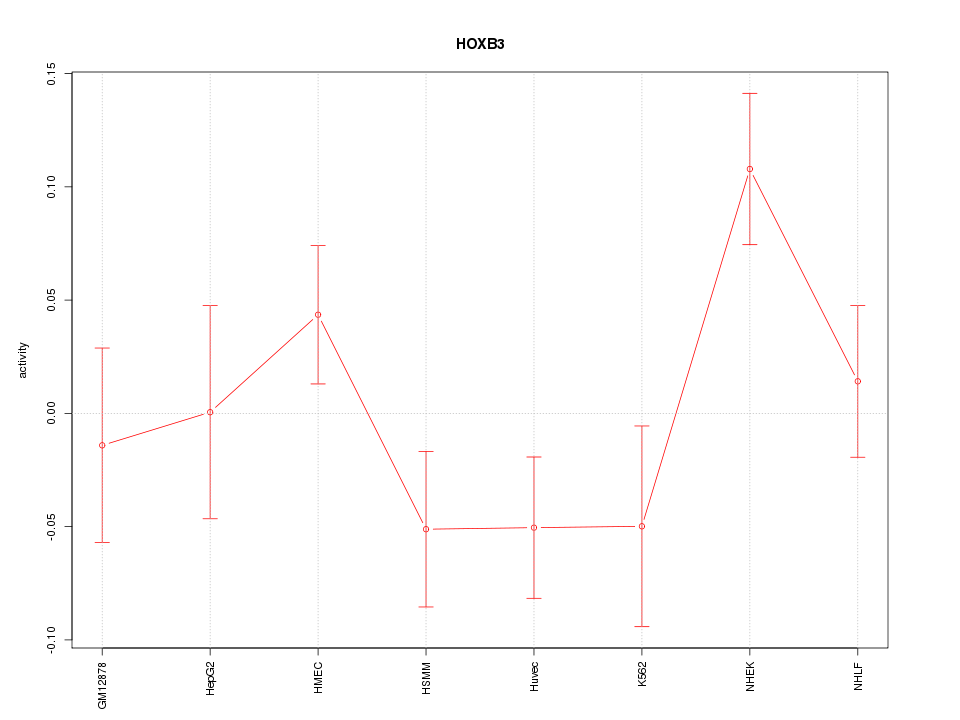
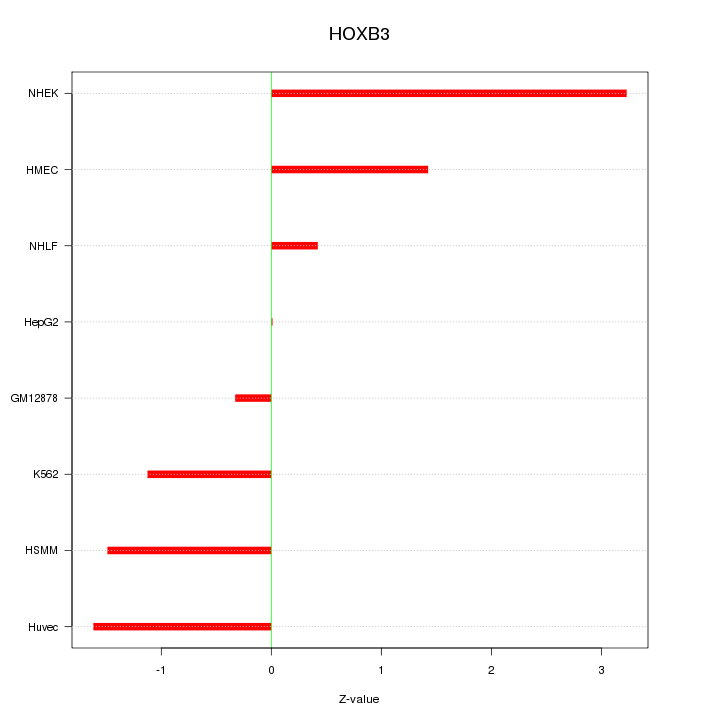
Motif ID: HOXB3
Z-value: 1.535
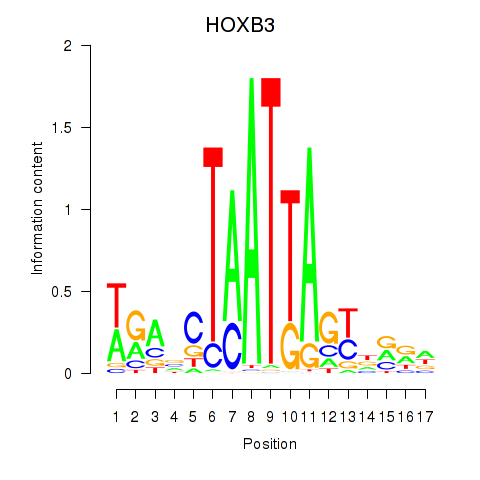
Transcription factors associated with HOXB3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HOXB3 | ENSG00000120093.7 | HOXB3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.8 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.8 | 2.4 | GO:0009635 | response to herbicide(GO:0009635) |
| 0.8 | 2.3 | GO:0014029 | neural crest formation(GO:0014029) neural crest cell fate commitment(GO:0014034) convergent extension involved in axis elongation(GO:0060028) cellular response to prostaglandin E stimulus(GO:0071380) response to heparin(GO:0071503) cellular response to heparin(GO:0071504) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.7 | 6.2 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.2 | 0.9 | GO:0031223 | auditory behavior(GO:0031223) malate-aspartate shuttle(GO:0043490) |
| 0.2 | 1.3 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.2 | 0.5 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) negative regulation of protein glycosylation(GO:0060051) |
| 0.2 | 1.8 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.2 | 1.4 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 | 1.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.4 | GO:1904037 | positive regulation of epithelial cell apoptotic process(GO:1904037) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.8 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.1 | 2.7 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.6 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.4 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.2 | GO:0042097 | lymphotoxin A production(GO:0032641) interleukin-4 biosynthetic process(GO:0042097) lymphotoxin A biosynthetic process(GO:0042109) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.2 | GO:0034587 | RNA 5'-end processing(GO:0000966) piRNA metabolic process(GO:0034587) |
| 0.1 | 0.2 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.2 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 1.8 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.6 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.1 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.0 | 4.0 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 1.1 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) |
| 0.0 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.5 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.6 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 1.0 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.0 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 1.4 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.1 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 1.1 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.3 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.1 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.8 | GO:0034249 | negative regulation of translation(GO:0017148) negative regulation of cellular amide metabolic process(GO:0034249) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 0.5 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 0.6 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 1.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.5 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 2.5 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.9 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.7 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 4.5 | GO:0005792 | obsolete microsome(GO:0005792) |
| 0.0 | 1.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 0.8 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.3 | 1.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 0.7 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.2 | 6.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 1.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 2.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 2.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.3 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 0.9 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) glutamate binding(GO:0016595) |
| 0.1 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.2 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 2.3 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 1.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 2.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.4 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 2.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.0 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.1 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.9 | SIG_BCR_SIGNALING_PATHWAY | Members of the BCR signaling pathway |