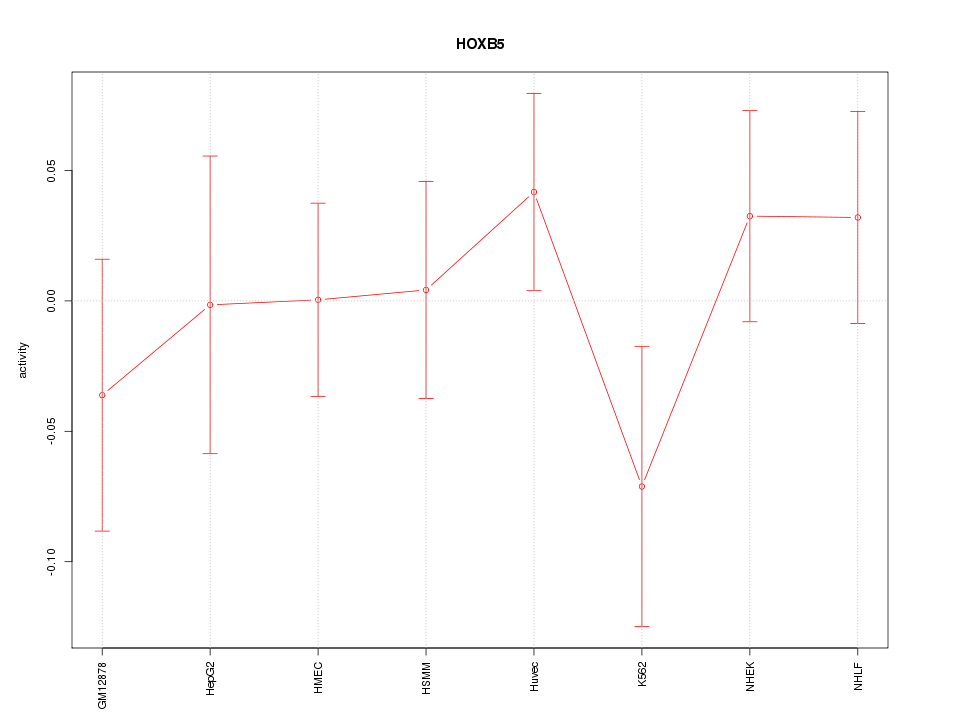
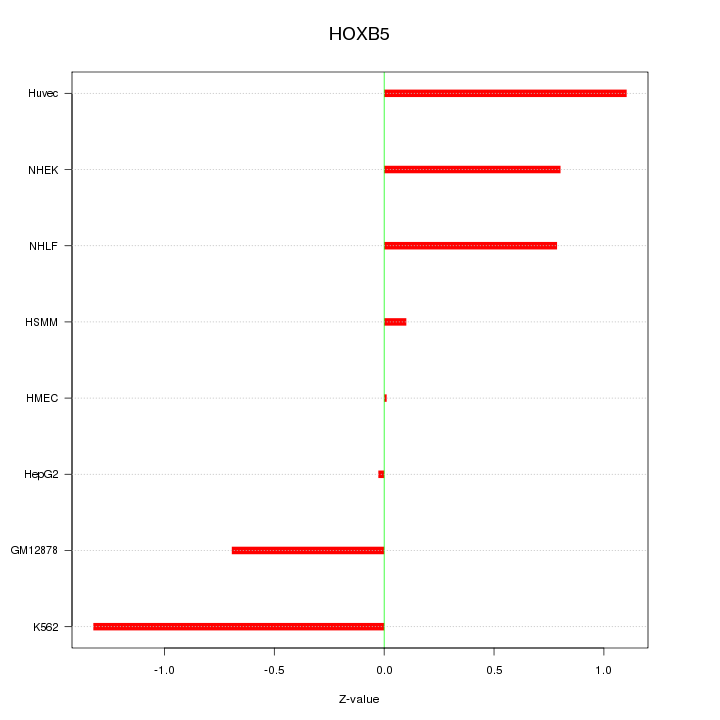
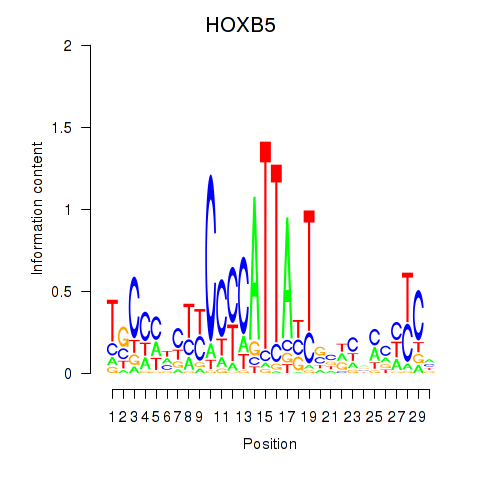
Motif ID: HOXB5
Z-value: 0.769
Transcription factors associated with HOXB5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HOXB5 | ENSG00000120075.5 | HOXB5 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0051503 | purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) |
| 0.2 | 0.7 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 1.3 | GO:2000124 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 0.5 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.1 | 1.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.4 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.1 | 0.2 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.1 | 0.2 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0032472 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.0 | 2.3 | GO:0030336 | negative regulation of cell migration(GO:0030336) negative regulation of cell motility(GO:2000146) |
| 0.0 | 0.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.8 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.5 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.1 | 1.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.4 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 1.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 2.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0046980 | oligopeptide-transporting ATPase activity(GO:0015421) tapasin binding(GO:0046980) |
| 0.0 | 0.2 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |