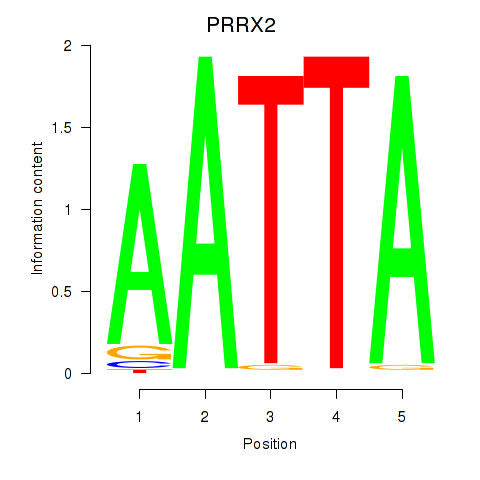
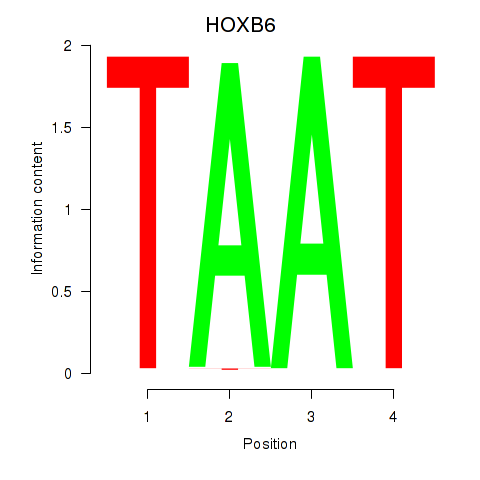
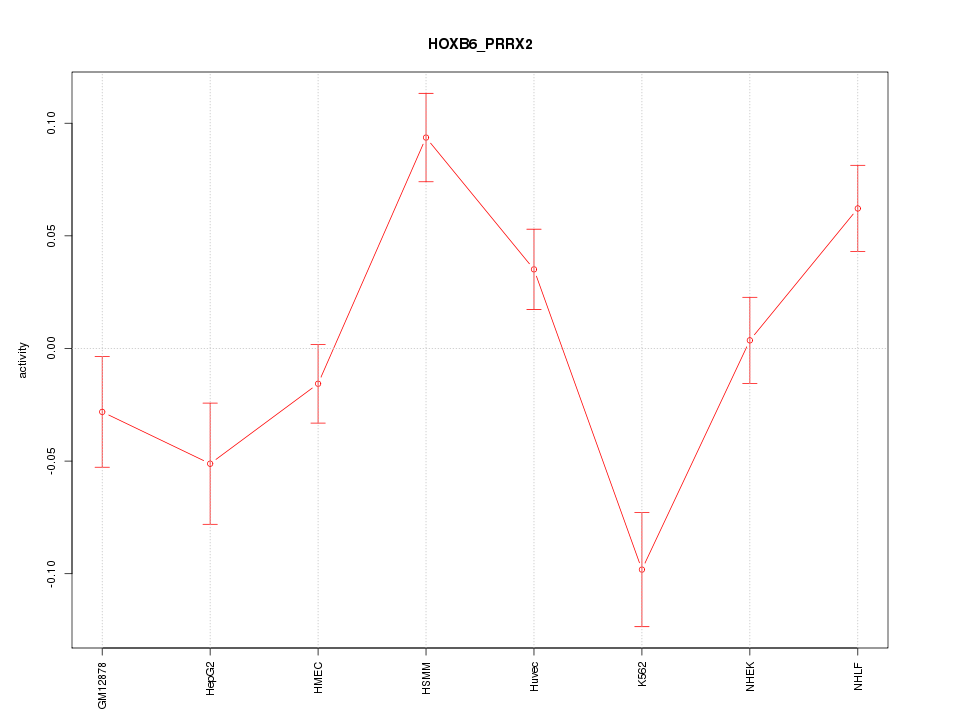
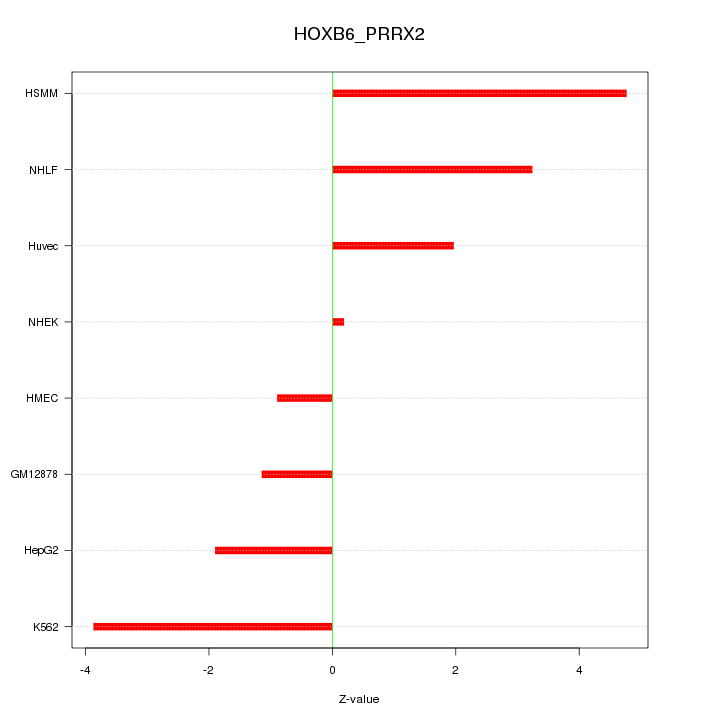
| 2.8 |
31.1 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.9 |
5.8 |
GO:2000121 |
regulation of removal of superoxide radicals(GO:2000121) |
| 1.4 |
2.8 |
GO:0060535 |
trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) |
| 1.4 |
4.1 |
GO:0007412 |
axon target recognition(GO:0007412) |
| 1.2 |
13.3 |
GO:0060216 |
definitive hemopoiesis(GO:0060216) |
| 1.1 |
3.2 |
GO:0014889 |
muscle atrophy(GO:0014889) regulation of skeletal muscle tissue growth(GO:0048631) |
| 1.0 |
3.1 |
GO:0060340 |
pericardium morphogenesis(GO:0003344) optic cup formation involved in camera-type eye development(GO:0003408) positive regulation of type I interferon-mediated signaling pathway(GO:0060340) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 1.0 |
8.2 |
GO:0042693 |
muscle cell fate commitment(GO:0042693) |
| 0.9 |
2.8 |
GO:0090047 |
obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.7 |
2.2 |
GO:0035987 |
endodermal cell fate commitment(GO:0001711) endodermal cell differentiation(GO:0035987) |
| 0.6 |
1.9 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.6 |
1.2 |
GO:0060585 |
regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.6 |
3.9 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.5 |
1.6 |
GO:0019731 |
antibacterial humoral response(GO:0019731) |
| 0.5 |
2.4 |
GO:0007182 |
common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.5 |
1.4 |
GO:0021836 |
chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.5 |
1.8 |
GO:0010716 |
regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.4 |
2.2 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.4 |
1.8 |
GO:0009956 |
radial pattern formation(GO:0009956) |
| 0.4 |
3.4 |
GO:0040037 |
negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.4 |
1.7 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.4 |
5.5 |
GO:0033687 |
osteoblast proliferation(GO:0033687) |
| 0.4 |
2.5 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.4 |
0.8 |
GO:0045617 |
negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.4 |
2.4 |
GO:0007499 |
ectoderm and mesoderm interaction(GO:0007499) |
| 0.4 |
2.3 |
GO:0060242 |
contact inhibition(GO:0060242) |
| 0.4 |
0.8 |
GO:0042701 |
progesterone secretion(GO:0042701) |
| 0.4 |
2.2 |
GO:2000124 |
cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.4 |
2.9 |
GO:0007351 |
tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.3 |
3.1 |
GO:0071692 |
sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.3 |
5.1 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.3 |
3.0 |
GO:0008354 |
germ cell migration(GO:0008354) |
| 0.3 |
0.7 |
GO:0010837 |
regulation of keratinocyte proliferation(GO:0010837) |
| 0.3 |
6.3 |
GO:0032012 |
regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 |
0.9 |
GO:0072677 |
eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.3 |
0.3 |
GO:0009441 |
glycolate metabolic process(GO:0009441) |
| 0.2 |
5.7 |
GO:0021846 |
cell proliferation in forebrain(GO:0021846) |
| 0.2 |
1.5 |
GO:0032472 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.2 |
0.5 |
GO:0033484 |
nitric oxide homeostasis(GO:0033484) |
| 0.2 |
2.9 |
GO:0060638 |
mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.2 |
0.7 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.2 |
1.4 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.2 |
1.8 |
GO:0030903 |
notochord development(GO:0030903) |
| 0.2 |
3.7 |
GO:0008347 |
glial cell migration(GO:0008347) |
| 0.2 |
0.6 |
GO:0048389 |
intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) trachea formation(GO:0060440) bud elongation involved in lung branching(GO:0060449) bud dilation involved in lung branching(GO:0060503) branch elongation involved in ureteric bud branching(GO:0060681) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) endothelial tube morphogenesis(GO:0061154) pulmonary artery endothelial tube morphogenesis(GO:0061155) pulmonary artery morphogenesis(GO:0061156) cell proliferation involved in mesonephros development(GO:0061209) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) negative regulation of mesonephros development(GO:0061218) pattern specification involved in mesonephros development(GO:0061227) renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) BMP signaling pathway involved in nephric duct formation(GO:0071893) glomerular visceral epithelial cell development(GO:0072015) comma-shaped body morphogenesis(GO:0072049) regulation of branch elongation involved in ureteric bud branching(GO:0072095) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) glomerulus morphogenesis(GO:0072102) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) nephric duct formation(GO:0072179) ureter urothelium development(GO:0072190) ureter epithelial cell differentiation(GO:0072192) ureter morphogenesis(GO:0072197) metanephric comma-shaped body morphogenesis(GO:0072278) glomerular epithelial cell development(GO:0072310) negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.2 |
1.1 |
GO:0002328 |
pro-B cell differentiation(GO:0002328) |
| 0.2 |
1.0 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.2 |
1.9 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 |
0.8 |
GO:1902116 |
negative regulation of organelle assembly(GO:1902116) |
| 0.2 |
0.6 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 |
0.4 |
GO:0035120 |
post-embryonic appendage morphogenesis(GO:0035120) |
| 0.2 |
1.1 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.2 |
0.6 |
GO:1903115 |
regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.2 |
1.3 |
GO:0032801 |
negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) receptor catabolic process(GO:0032801) regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) |
| 0.2 |
22.8 |
GO:0006367 |
transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.2 |
1.5 |
GO:0021952 |
central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 |
4.9 |
GO:0034199 |
activation of protein kinase A activity(GO:0034199) |
| 0.2 |
1.1 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.2 |
6.7 |
GO:0090263 |
positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.2 |
0.9 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 |
0.6 |
GO:0045880 |
positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.2 |
3.9 |
GO:0008038 |
neuron recognition(GO:0008038) |
| 0.2 |
3.1 |
GO:0045446 |
endothelium development(GO:0003158) endothelial cell differentiation(GO:0045446) |
| 0.2 |
2.7 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.1 |
0.4 |
GO:0001941 |
postsynaptic membrane organization(GO:0001941) |
| 0.1 |
1.2 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 |
0.9 |
GO:0042730 |
fibrinolysis(GO:0042730) |
| 0.1 |
1.9 |
GO:0050872 |
white fat cell differentiation(GO:0050872) |
| 0.1 |
0.6 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.1 |
0.5 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 |
3.1 |
GO:0006555 |
methionine metabolic process(GO:0006555) |
| 0.1 |
2.6 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.1 |
1.7 |
GO:0000052 |
citrulline metabolic process(GO:0000052) |
| 0.1 |
3.2 |
GO:0017158 |
regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.1 |
0.2 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 |
0.9 |
GO:0003065 |
regulation of heart rate by chemical signal(GO:0003062) positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 |
0.6 |
GO:0042474 |
middle ear morphogenesis(GO:0042474) |
| 0.1 |
1.1 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 |
0.1 |
GO:0010982 |
regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 |
0.3 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.1 |
7.2 |
GO:0030049 |
muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 |
0.3 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.1 |
0.4 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
4.2 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 |
1.3 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
1.3 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.1 |
0.1 |
GO:0070857 |
regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.1 |
1.0 |
GO:0031641 |
regulation of myelination(GO:0031641) |
| 0.1 |
4.2 |
GO:0007585 |
respiratory gaseous exchange(GO:0007585) |
| 0.1 |
1.7 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.1 |
0.5 |
GO:0055062 |
phosphate ion homeostasis(GO:0055062) divalent inorganic anion homeostasis(GO:0072505) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.1 |
18.7 |
GO:0007517 |
muscle organ development(GO:0007517) |
| 0.1 |
2.2 |
GO:0007368 |
determination of left/right symmetry(GO:0007368) |
| 0.1 |
0.6 |
GO:0071320 |
cellular response to cAMP(GO:0071320) |
| 0.1 |
3.0 |
GO:0051101 |
regulation of DNA binding(GO:0051101) |
| 0.1 |
1.0 |
GO:1901797 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.1 |
0.2 |
GO:0045760 |
positive regulation of action potential(GO:0045760) |
| 0.1 |
0.6 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.1 |
0.1 |
GO:0010658 |
striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 |
0.7 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 |
6.0 |
GO:0007229 |
integrin-mediated signaling pathway(GO:0007229) |
| 0.1 |
0.5 |
GO:0032528 |
microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 |
0.8 |
GO:0031532 |
actin cytoskeleton reorganization(GO:0031532) |
| 0.1 |
2.8 |
GO:0060271 |
cilium morphogenesis(GO:0060271) |
| 0.1 |
3.9 |
GO:0009408 |
response to heat(GO:0009408) |
| 0.1 |
2.5 |
GO:0008016 |
regulation of heart contraction(GO:0008016) |
| 0.1 |
0.7 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.1 |
2.6 |
GO:0060828 |
regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.1 |
0.2 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.1 |
0.6 |
GO:0007528 |
neuromuscular junction development(GO:0007528) |
| 0.1 |
0.3 |
GO:0009756 |
carbohydrate mediated signaling(GO:0009756) |
| 0.1 |
0.4 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.1 |
0.3 |
GO:0097531 |
mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 |
3.1 |
GO:0031032 |
actomyosin structure organization(GO:0031032) |
| 0.1 |
0.6 |
GO:0030853 |
negative regulation of granulocyte differentiation(GO:0030853) |
| 0.1 |
0.3 |
GO:0051963 |
regulation of synapse assembly(GO:0051963) |
| 0.0 |
0.2 |
GO:0006848 |
pyruvate transport(GO:0006848) |
| 0.0 |
0.9 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.0 |
0.4 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
0.1 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 |
1.1 |
GO:0050850 |
positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.0 |
0.2 |
GO:0032488 |
Cdc42 protein signal transduction(GO:0032488) |
| 0.0 |
0.5 |
GO:0006907 |
pinocytosis(GO:0006907) |
| 0.0 |
0.6 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.0 |
0.1 |
GO:0009299 |
mRNA transcription(GO:0009299) |
| 0.0 |
0.5 |
GO:0051654 |
establishment of mitochondrion localization(GO:0051654) |
| 0.0 |
8.4 |
GO:0006936 |
muscle contraction(GO:0006936) |
| 0.0 |
2.3 |
GO:0030198 |
extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 |
1.0 |
GO:0051180 |
vitamin transport(GO:0051180) |
| 0.0 |
1.5 |
GO:0048813 |
dendrite morphogenesis(GO:0048813) |
| 0.0 |
0.1 |
GO:0071460 |
cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 |
0.7 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.8 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.5 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.0 |
0.2 |
GO:0007635 |
chemosensory behavior(GO:0007635) |
| 0.0 |
0.1 |
GO:0008628 |
hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 |
0.2 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.3 |
GO:0006184 |
obsolete GTP catabolic process(GO:0006184) |
| 0.0 |
0.3 |
GO:0035315 |
hair cell differentiation(GO:0035315) auditory receptor cell differentiation(GO:0042491) |
| 0.0 |
0.5 |
GO:0045773 |
positive regulation of axon extension(GO:0045773) |
| 0.0 |
0.8 |
GO:0048199 |
vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 |
0.1 |
GO:0001569 |
patterning of blood vessels(GO:0001569) |
| 0.0 |
2.2 |
GO:0009952 |
anterior/posterior pattern specification(GO:0009952) |
| 0.0 |
0.2 |
GO:0009082 |
branched-chain amino acid biosynthetic process(GO:0009082) |
| 0.0 |
0.2 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.3 |
GO:0015914 |
phospholipid transport(GO:0015914) |
| 0.0 |
0.7 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
0.2 |
GO:0044245 |
polysaccharide digestion(GO:0044245) |
| 0.0 |
0.1 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 |
0.1 |
GO:0090083 |
regulation of inclusion body assembly(GO:0090083) |
| 0.0 |
0.3 |
GO:0008272 |
sulfate transport(GO:0008272) |
| 0.0 |
0.5 |
GO:0042776 |
mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 |
1.4 |
GO:0006754 |
ATP biosynthetic process(GO:0006754) |
| 0.0 |
2.7 |
GO:0043087 |
regulation of GTPase activity(GO:0043087) |
| 0.0 |
0.1 |
GO:0006216 |
cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 |
1.6 |
GO:0007265 |
Ras protein signal transduction(GO:0007265) |
| 0.0 |
1.4 |
GO:0070304 |
positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
| 0.0 |
0.1 |
GO:0045066 |
regulatory T cell differentiation(GO:0045066) |
| 0.0 |
3.0 |
GO:0030036 |
actin cytoskeleton organization(GO:0030036) |
| 0.0 |
2.8 |
GO:0050658 |
nucleic acid transport(GO:0050657) RNA transport(GO:0050658) establishment of RNA localization(GO:0051236) |
| 0.0 |
0.3 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.4 |
GO:0006368 |
transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 |
1.8 |
GO:0008203 |
cholesterol metabolic process(GO:0008203) |
| 0.0 |
0.1 |
GO:0044253 |
positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 |
0.2 |
GO:1901224 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 |
0.2 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 |
3.7 |
GO:0006260 |
DNA replication(GO:0006260) |
| 0.0 |
1.0 |
GO:0006986 |
response to unfolded protein(GO:0006986) |
| 0.0 |
0.2 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.1 |
GO:0046174 |
polyol catabolic process(GO:0046174) |
| 0.0 |
0.3 |
GO:0043631 |
RNA polyadenylation(GO:0043631) |
| 0.0 |
0.1 |
GO:0032230 |
positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.0 |
0.1 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 |
0.4 |
GO:0030866 |
cortical cytoskeleton organization(GO:0030865) cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
5.1 |
GO:0016567 |
protein ubiquitination(GO:0016567) |
| 0.0 |
0.4 |
GO:0019722 |
calcium-mediated signaling(GO:0019722) |
| 0.0 |
0.1 |
GO:0035115 |
embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 |
0.1 |
GO:0051319 |
mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 |
2.8 |
GO:0051056 |
regulation of small GTPase mediated signal transduction(GO:0051056) |