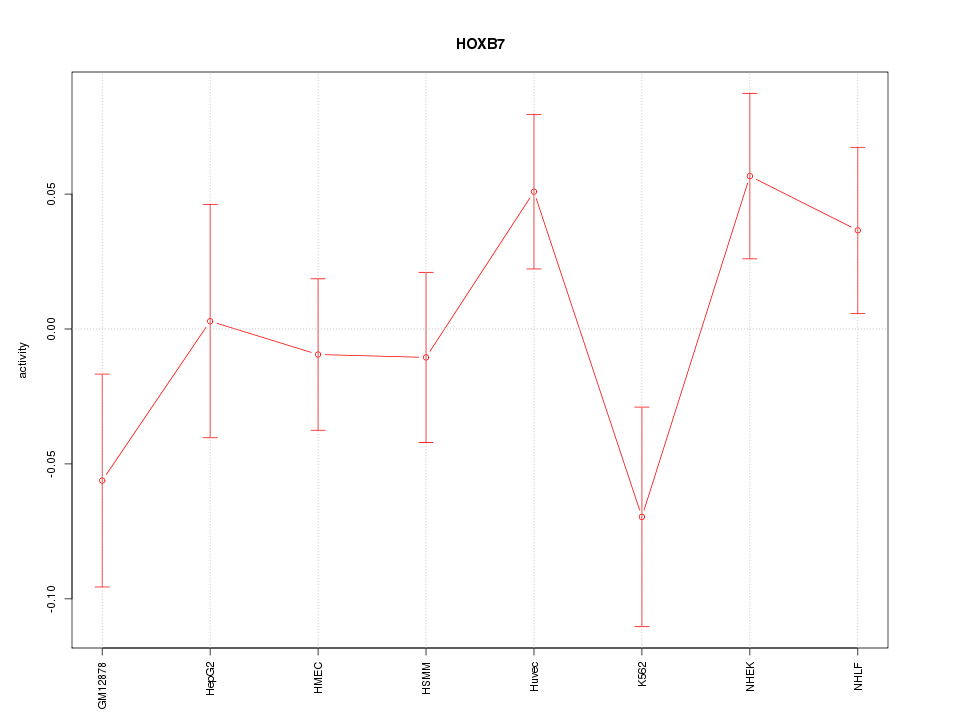
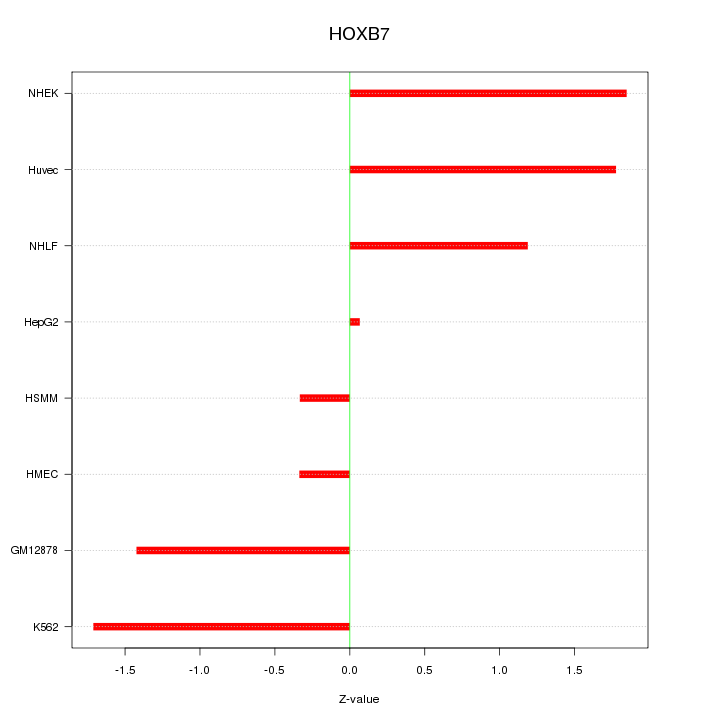
Motif ID: HOXB7
Z-value: 1.283
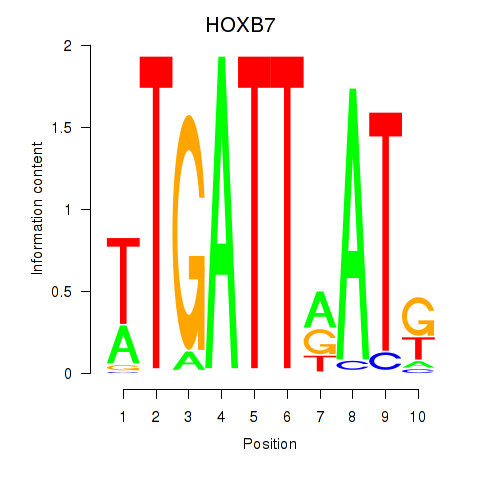
Transcription factors associated with HOXB7:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HOXB7 | ENSG00000260027.3 | HOXB7 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.8 | 4.2 | GO:0006789 | bilirubin conjugation(GO:0006789) biphenyl metabolic process(GO:0018879) biphenyl catabolic process(GO:0070980) |
| 0.6 | 2.9 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.4 | 1.8 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.4 | 1.5 | GO:0060649 | nipple development(GO:0060618) mammary gland bud elongation(GO:0060649) nipple morphogenesis(GO:0060658) nipple sheath formation(GO:0060659) |
| 0.3 | 0.8 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.3 | 2.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 1.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 2.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 1.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 0.7 | GO:0001302 | replicative cell aging(GO:0001302) cloaca development(GO:0035844) female genitalia morphogenesis(GO:0048807) cloacal septation(GO:0060197) |
| 0.1 | 1.9 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.1 | 0.8 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.1 | 0.5 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 | 0.4 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.4 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.8 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.1 | 2.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.6 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.1 | 1.1 | GO:0032727 | positive regulation of interferon-alpha production(GO:0032727) |
| 0.1 | 0.3 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.1 | 0.1 | GO:2000831 | corticosteroid hormone secretion(GO:0035930) regulation of steroid hormone secretion(GO:2000831) regulation of corticosteroid hormone secretion(GO:2000846) |
| 0.1 | 2.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 1.0 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.1 | 0.5 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 1.0 | GO:0010658 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 0.2 | GO:0048861 | serine phosphorylation of STAT protein(GO:0042501) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.4 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 0.6 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.4 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.1 | 1.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.3 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 1.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0035283 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.5 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.8 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.0 | 0.3 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.2 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.9 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.4 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.9 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:0071801 | epithelial fluid transport(GO:0042045) podosome assembly(GO:0071800) regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.8 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.2 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 1.2 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.2 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.3 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.3 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 | 0.5 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.0 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.4 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.4 | GO:0006693 | prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.2 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.7 | GO:0045727 | positive regulation of cellular amide metabolic process(GO:0034250) positive regulation of translation(GO:0045727) |
| 0.0 | 0.2 | GO:0042436 | tryptophan metabolic process(GO:0006568) tryptophan catabolic process(GO:0006569) indolalkylamine metabolic process(GO:0006586) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 0.9 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.2 | 2.8 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.7 | GO:0019718 | obsolete rough microsome(GO:0019718) |
| 0.1 | 1.7 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 6.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 3.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.6 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 8.7 | GO:0005792 | obsolete microsome(GO:0005792) |
| 0.0 | 1.9 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 5.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.5 | 1.9 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.4 | 1.1 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.3 | 8.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 2.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 0.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 0.7 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.4 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.1 | 0.4 | GO:0047718 | indanol dehydrogenase activity(GO:0047718) |
| 0.1 | 0.8 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.8 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.3 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 0.8 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 2.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 1.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.2 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 0.8 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.2 | GO:0004769 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 2.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.4 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 1.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 2.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.2 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0017096 | acetylserotonin O-methyltransferase activity(GO:0017096) |
| 0.0 | 1.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 5.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.2 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.0 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.0 | 0.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.9 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 2.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | SIG_BCR_SIGNALING_PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.6 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |