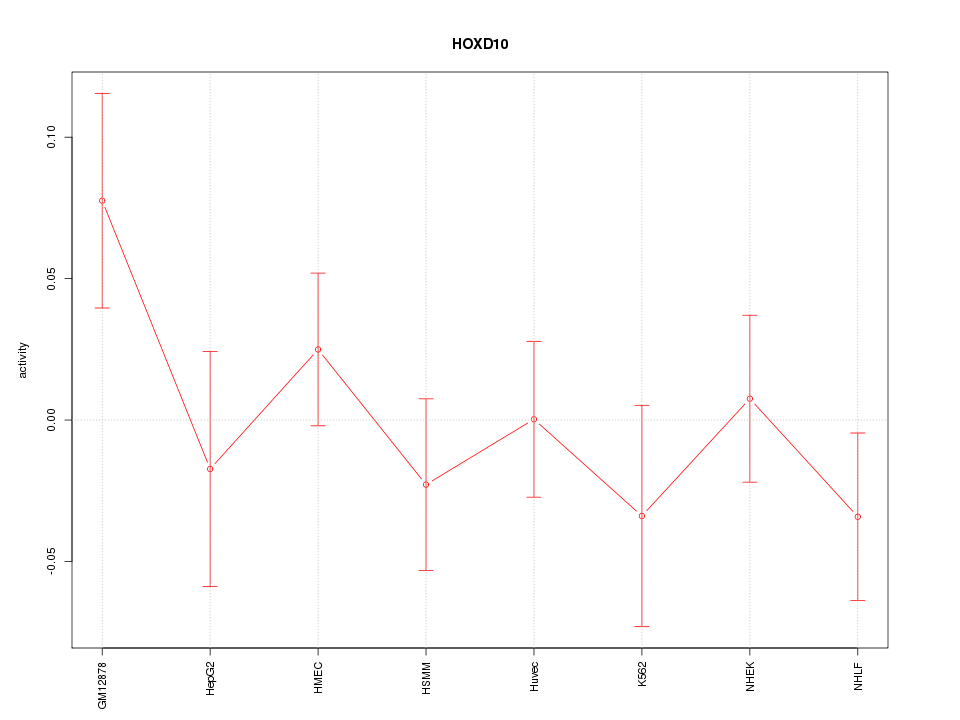
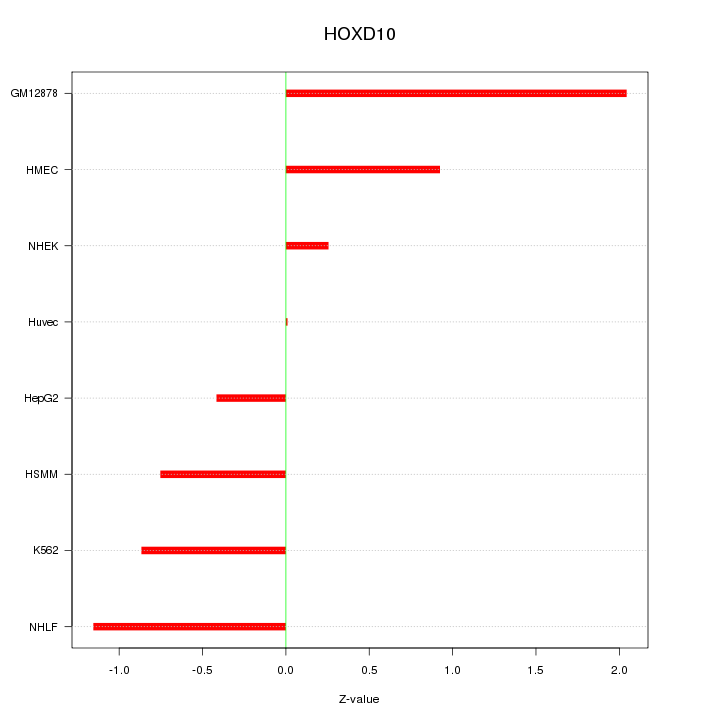
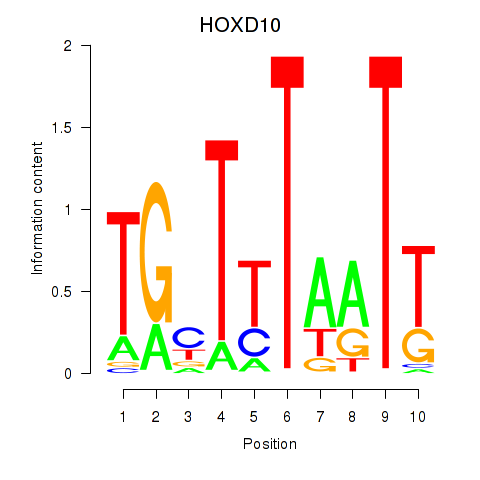
Motif ID: HOXD10
Z-value: 0.995
Transcription factors associated with HOXD10:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HOXD10 | ENSG00000128710.5 | HOXD10 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.2 | 0.8 | GO:0070527 | integrin activation(GO:0033622) regulation of integrin activation(GO:0033623) positive regulation of integrin activation(GO:0033625) platelet aggregation(GO:0070527) |
| 0.2 | 5.0 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.2 | 0.6 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.6 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 0.4 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) positive regulation of vesicle fusion(GO:0031340) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.3 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.1 | 0.2 | GO:0001552 | ovarian follicle atresia(GO:0001552) |
| 0.1 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.3 | GO:0043316 | natural killer cell activation involved in immune response(GO:0002323) cytotoxic T cell degranulation(GO:0043316) natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.3 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.2 | GO:0021896 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.1 | 0.3 | GO:0032472 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.1 | 1.2 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.6 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.2 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 1.4 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.6 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 1.7 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.8 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.8 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.3 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.0 | 0.2 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.2 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.1 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.0 | 0.3 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.1 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 1.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 2.6 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.2 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.0 | GO:0010430 | ethanol catabolic process(GO:0006068) fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.2 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.3 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 1.6 | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.1 | GO:0032099 | negative regulation of appetite(GO:0032099) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 0.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 1.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) fascia adherens(GO:0005916) |
| 0.0 | 0.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) synaptic cleft(GO:0043083) cell body membrane(GO:0044298) |
| 0.0 | 0.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 2.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.6 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.3 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 1.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 1.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.3 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.6 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.0 | 0.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 1.0 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 1.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:0050833 | pyruvate secondary active transmembrane transporter activity(GO:0005477) pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.0 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 2.4 | GO:0003702 | obsolete RNA polymerase II transcription factor activity(GO:0003702) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.0 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |