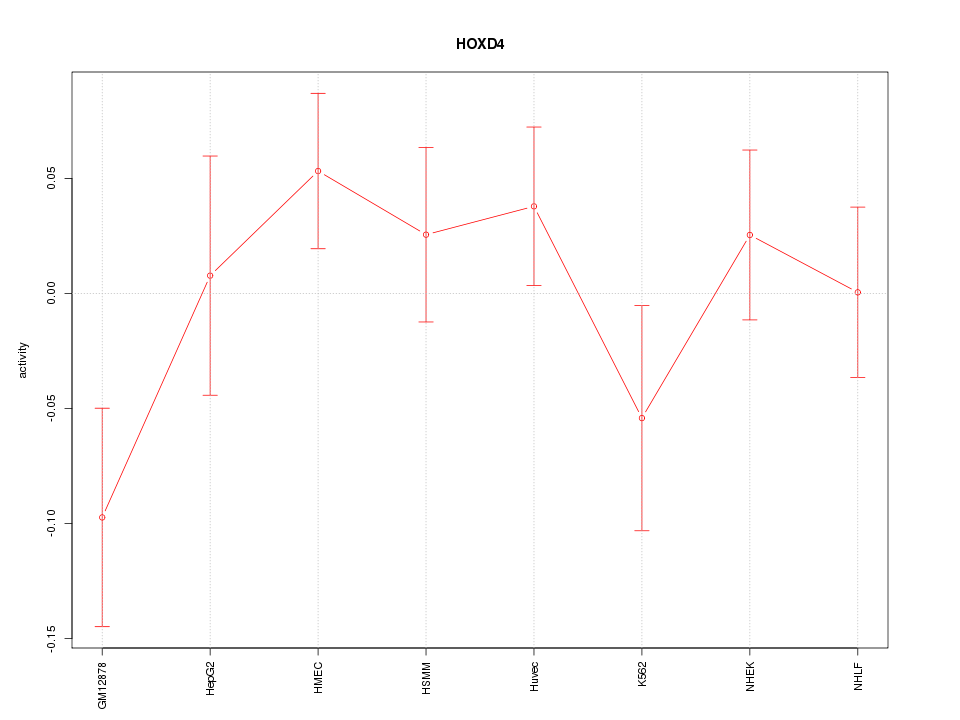
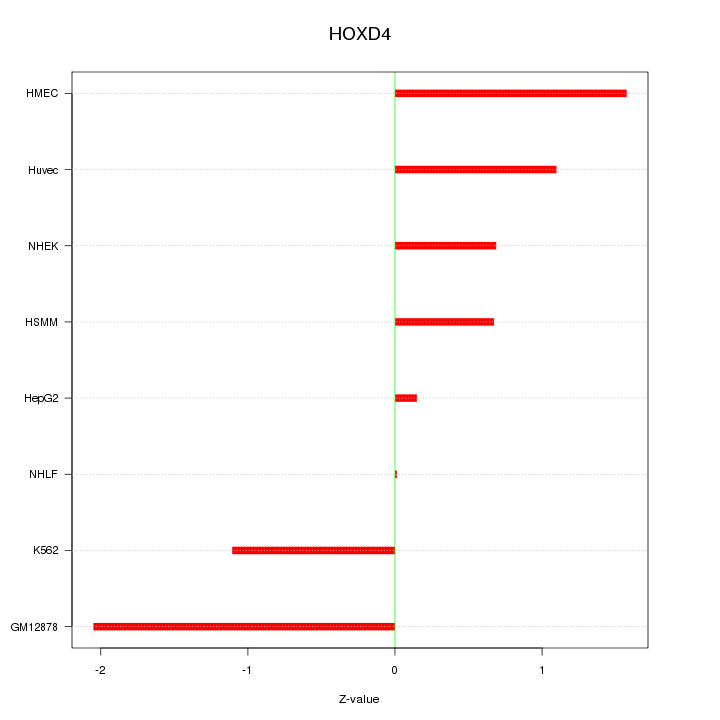
Motif ID: HOXD4
Z-value: 1.122
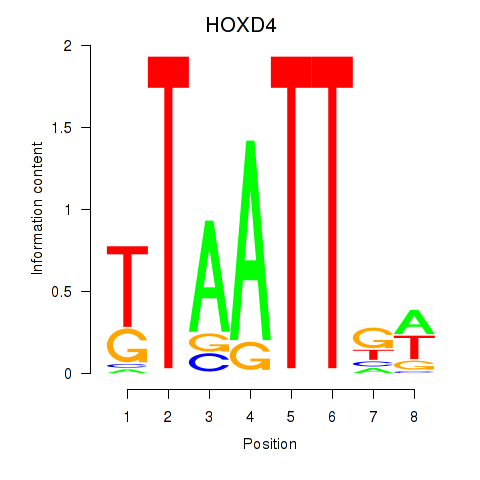
Transcription factors associated with HOXD4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HOXD4 | ENSG00000170166.5 | HOXD4 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) positive regulation of vesicle fusion(GO:0031340) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.3 | 1.6 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.2 | 1.4 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 | 0.8 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.4 | GO:0014889 | muscle atrophy(GO:0014889) regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 | 0.5 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.1 | 0.4 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.1 | 2.1 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.5 | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) |
| 0.1 | 0.5 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.5 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 0.2 | GO:0046621 | negative regulation of organ growth(GO:0046621) |
| 0.1 | 0.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 1.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 2.0 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) |
| 0.0 | 0.9 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.8 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 1.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.2 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.2 | GO:0046051 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.0 | GO:0019056 | modulation by virus of host transcription(GO:0019056) regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 1.8 | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.0 | 2.2 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.2 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 4.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.3 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 2.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.8 | 3.9 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 2.1 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.2 | 0.5 | GO:0050816 | beta-2 adrenergic receptor binding(GO:0031698) phosphothreonine binding(GO:0050816) |
| 0.1 | 1.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.3 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 0.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 1.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.7 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 1.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |