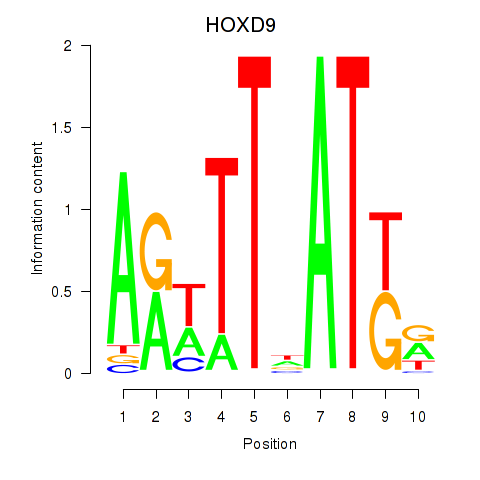
Motif ID: HOXD9
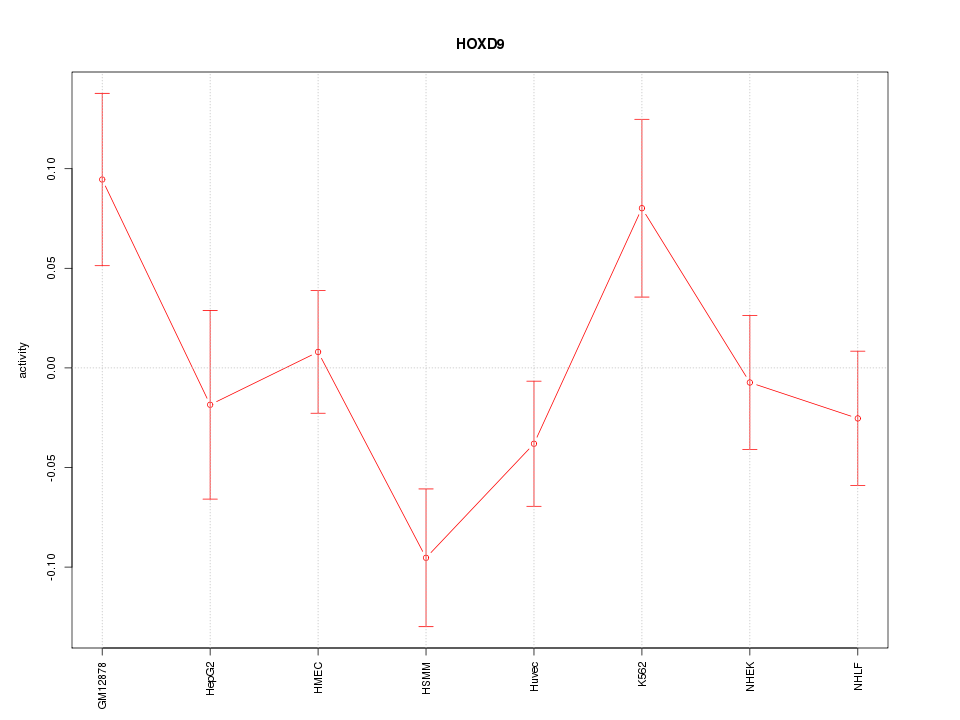
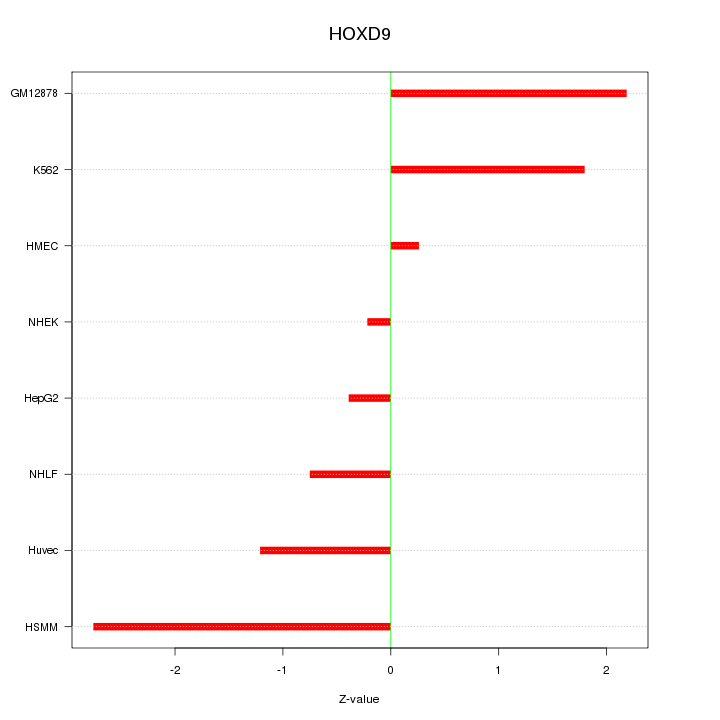
Z-value: 1.496
Transcription factors associated with HOXD9:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HOXD9 | ENSG00000128709.10 | HOXD9 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.7 | 2.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.3 | 1.5 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.3 | 1.7 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.2 | 1.3 | GO:0043316 | natural killer cell activation involved in immune response(GO:0002323) cytotoxic T cell degranulation(GO:0043316) natural killer cell degranulation(GO:0043320) |
| 0.2 | 0.6 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 0.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.3 | GO:0033364 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.5 | GO:0030578 | nuclear body organization(GO:0030575) PML body organization(GO:0030578) |
| 0.1 | 0.7 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.4 | GO:1904037 | positive regulation of epithelial cell apoptotic process(GO:1904037) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.3 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.1 | 0.3 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.7 | GO:0006863 | purine nucleobase transport(GO:0006863) |
| 0.1 | 0.3 | GO:0031033 | positive regulation of lamellipodium assembly(GO:0010592) myosin filament organization(GO:0031033) positive regulation of lamellipodium organization(GO:1902745) |
| 0.1 | 0.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.3 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) positive regulation of activation of membrane attack complex(GO:0001970) positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.1 | 0.4 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 1.3 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.6 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 1.9 | GO:0000080 | mitotic G1 phase(GO:0000080) |
| 0.0 | 0.2 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 2.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.3 | GO:0045822 | negative regulation of heart contraction(GO:0045822) |
| 0.0 | 0.6 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.4 | GO:0006807 | nitrogen compound metabolic process(GO:0006807) |
| 0.0 | 0.9 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.0 | 0.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 1.0 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 2.2 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 0.1 | GO:0042519 | tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.0 | 2.3 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 0.6 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 2.5 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 1.2 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.0 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.2 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 1.5 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 0.4 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 1.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.4 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 1.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.7 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.3 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 1.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.4 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 2.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.1 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.6 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.3 | 1.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 2.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 0.7 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 1.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.4 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 1.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.7 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) |
| 0.1 | 1.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.3 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 2.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 1.7 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.9 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 2.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 1.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.3 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0060590 | ATPase regulator activity(GO:0060590) |
| 0.0 | 0.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.3 | GO:0043560 | epidermal growth factor receptor binding(GO:0005154) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 1.4 | GO:0005178 | integrin binding(GO:0005178) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.0 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |