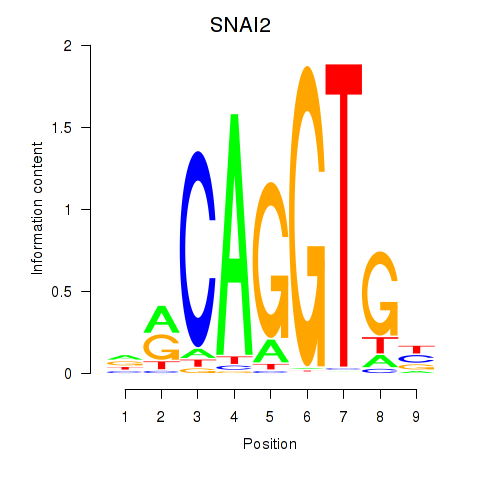
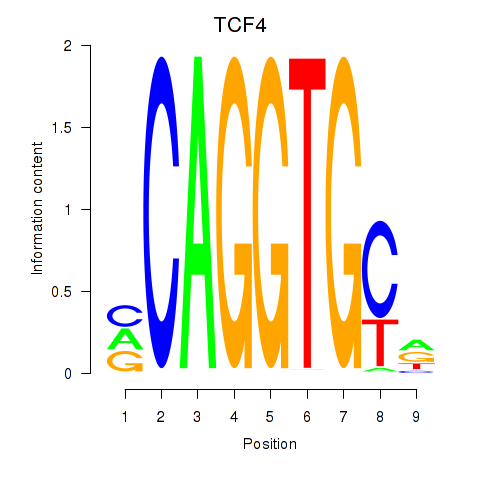
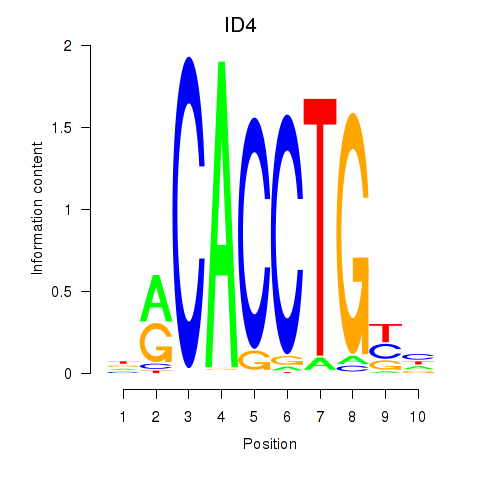
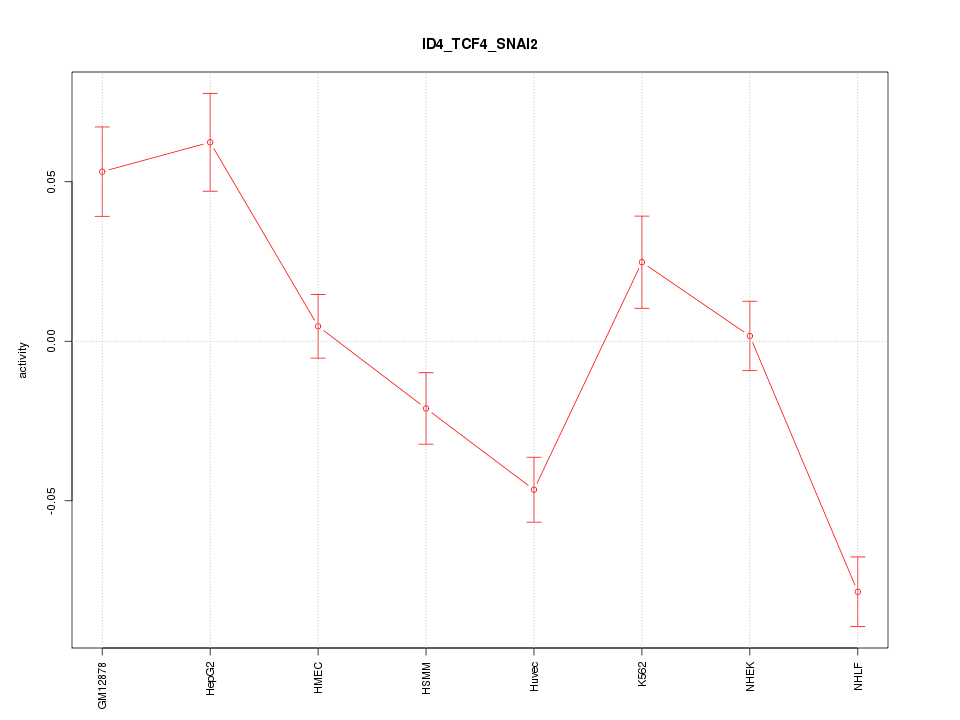
| 2.2 |
6.6 |
GO:0051066 |
dihydrobiopterin metabolic process(GO:0051066) |
| 1.9 |
5.8 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 1.2 |
3.7 |
GO:0061298 |
retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 1.2 |
3.5 |
GO:2000910 |
regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 1.1 |
3.2 |
GO:0000738 |
DNA catabolic process, exonucleolytic(GO:0000738) |
| 1.0 |
3.9 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.9 |
3.6 |
GO:0032291 |
central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.8 |
3.3 |
GO:0010900 |
regulation of phosphatidylcholine catabolic process(GO:0010899) negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.8 |
3.3 |
GO:0072180 |
mesonephric duct morphogenesis(GO:0072180) |
| 0.8 |
3.3 |
GO:0071364 |
cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.8 |
4.9 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.8 |
3.2 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.8 |
2.4 |
GO:0060696 |
regulation of phospholipid catabolic process(GO:0060696) |
| 0.8 |
5.4 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.7 |
2.2 |
GO:0031077 |
post-embryonic camera-type eye development(GO:0031077) Spemann organizer formation(GO:0060061) |
| 0.7 |
3.7 |
GO:0009439 |
cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.7 |
0.7 |
GO:0006930 |
substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.7 |
2.1 |
GO:0032328 |
alanine transport(GO:0032328) |
| 0.7 |
0.7 |
GO:0070874 |
negative regulation of glycogen metabolic process(GO:0070874) |
| 0.7 |
2.1 |
GO:0090206 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.7 |
2.1 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.6 |
2.5 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.6 |
0.6 |
GO:0042403 |
thyroid hormone metabolic process(GO:0042403) |
| 0.6 |
1.8 |
GO:0032928 |
regulation of superoxide anion generation(GO:0032928) negative regulation of superoxide anion generation(GO:0032929) |
| 0.6 |
1.8 |
GO:0043366 |
beta selection(GO:0043366) |
| 0.6 |
2.4 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.6 |
0.6 |
GO:0043049 |
otic placode formation(GO:0043049) |
| 0.6 |
1.8 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.6 |
1.8 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.6 |
5.0 |
GO:0006546 |
glycine catabolic process(GO:0006546) |
| 0.5 |
1.6 |
GO:0006768 |
biotin metabolic process(GO:0006768) |
| 0.5 |
1.6 |
GO:0009107 |
lipoate biosynthetic process(GO:0009107) |
| 0.5 |
1.6 |
GO:0006929 |
substrate-dependent cell migration(GO:0006929) |
| 0.5 |
1.6 |
GO:0019886 |
antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.5 |
0.5 |
GO:0033622 |
integrin activation(GO:0033622) regulation of integrin activation(GO:0033623) positive regulation of integrin activation(GO:0033625) |
| 0.5 |
2.1 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.5 |
2.6 |
GO:0032878 |
regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.5 |
5.6 |
GO:0098927 |
early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.5 |
1.5 |
GO:1904742 |
regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) regulation of telomeric DNA binding(GO:1904742) positive regulation of telomeric DNA binding(GO:1904744) |
| 0.5 |
1.5 |
GO:0043095 |
regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.5 |
1.5 |
GO:0071603 |
endothelial cell-cell adhesion(GO:0071603) |
| 0.5 |
1.5 |
GO:0046210 |
nitric oxide catabolic process(GO:0046210) |
| 0.5 |
1.4 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.5 |
1.0 |
GO:0014874 |
response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.5 |
1.4 |
GO:0032571 |
response to vitamin K(GO:0032571) |
| 0.5 |
2.7 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.5 |
1.4 |
GO:0006552 |
leucine catabolic process(GO:0006552) |
| 0.4 |
1.8 |
GO:0010727 |
negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.4 |
1.7 |
GO:0002478 |
antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 0.4 |
4.7 |
GO:0046628 |
positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.4 |
1.3 |
GO:0035408 |
histone H3-T6 phosphorylation(GO:0035408) |
| 0.4 |
1.7 |
GO:0042816 |
pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.4 |
2.1 |
GO:0003070 |
age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) |
| 0.4 |
2.5 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.4 |
3.7 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.4 |
1.6 |
GO:0009609 |
response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.4 |
3.3 |
GO:0019388 |
galactose catabolic process(GO:0019388) |
| 0.4 |
0.8 |
GO:0055078 |
sodium ion homeostasis(GO:0055078) |
| 0.4 |
1.2 |
GO:0046010 |
positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.4 |
1.9 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.4 |
1.5 |
GO:0015917 |
aminophospholipid transport(GO:0015917) |
| 0.4 |
1.1 |
GO:0070340 |
detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.4 |
1.9 |
GO:0051450 |
myoblast proliferation(GO:0051450) |
| 0.4 |
0.7 |
GO:0002439 |
chronic inflammatory response to antigenic stimulus(GO:0002439) positive regulation of chronic inflammatory response(GO:0002678) regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.4 |
1.1 |
GO:0009635 |
response to herbicide(GO:0009635) |
| 0.4 |
0.7 |
GO:0042632 |
cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.4 |
0.7 |
GO:0002677 |
negative regulation of chronic inflammatory response(GO:0002677) |
| 0.4 |
1.4 |
GO:0034379 |
very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.3 |
1.0 |
GO:0045656 |
negative regulation of monocyte differentiation(GO:0045656) |
| 0.3 |
3.1 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.3 |
1.0 |
GO:0034635 |
glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.3 |
2.0 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.3 |
1.6 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.3 |
1.3 |
GO:0051712 |
positive regulation of killing of cells of other organism(GO:0051712) |
| 0.3 |
1.3 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 |
1.3 |
GO:0010944 |
negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.3 |
2.2 |
GO:0033600 |
negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.3 |
2.2 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.3 |
4.4 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.3 |
0.6 |
GO:0032898 |
neurotrophin production(GO:0032898) |
| 0.3 |
6.2 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.3 |
1.2 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.3 |
7.1 |
GO:0046627 |
negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.3 |
1.8 |
GO:0009450 |
gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.3 |
2.7 |
GO:0016998 |
cell wall macromolecule catabolic process(GO:0016998) |
| 0.3 |
1.2 |
GO:0031580 |
membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.3 |
0.6 |
GO:0060896 |
neural plate anterior/posterior regionalization(GO:0021999) neural plate pattern specification(GO:0060896) neural plate regionalization(GO:0060897) |
| 0.3 |
0.6 |
GO:0045956 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.3 |
0.9 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.3 |
8.5 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.3 |
0.9 |
GO:0051781 |
positive regulation of cell division(GO:0051781) |
| 0.3 |
0.8 |
GO:0045978 |
regulation of oxidative phosphorylation(GO:0002082) negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.3 |
0.6 |
GO:0032119 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.3 |
1.6 |
GO:0046951 |
ketone body biosynthetic process(GO:0046951) |
| 0.3 |
0.8 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.3 |
0.3 |
GO:0051770 |
positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.3 |
0.3 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.3 |
0.8 |
GO:0048627 |
myoblast development(GO:0048627) |
| 0.3 |
1.9 |
GO:0090400 |
stress-induced premature senescence(GO:0090400) |
| 0.3 |
2.4 |
GO:0030277 |
epithelial structure maintenance(GO:0010669) maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.3 |
3.4 |
GO:0031498 |
nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.3 |
0.8 |
GO:0052805 |
histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.3 |
1.8 |
GO:0015755 |
fructose transport(GO:0015755) |
| 0.3 |
0.5 |
GO:0001942 |
hair follicle development(GO:0001942) molting cycle process(GO:0022404) hair cycle process(GO:0022405) molting cycle(GO:0042303) hair cycle(GO:0042633) skin epidermis development(GO:0098773) |
| 0.3 |
0.5 |
GO:0003176 |
aortic valve development(GO:0003176) aortic valve morphogenesis(GO:0003180) |
| 0.3 |
2.3 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.3 |
2.0 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.3 |
1.3 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.2 |
0.7 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.2 |
2.0 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.2 |
1.7 |
GO:0002360 |
T cell lineage commitment(GO:0002360) |
| 0.2 |
0.7 |
GO:0046292 |
formaldehyde metabolic process(GO:0046292) |
| 0.2 |
2.4 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.2 |
0.5 |
GO:0001977 |
renal system process involved in regulation of blood volume(GO:0001977) |
| 0.2 |
0.2 |
GO:0043173 |
purine nucleotide salvage(GO:0032261) nucleotide salvage(GO:0043173) |
| 0.2 |
1.4 |
GO:0051973 |
positive regulation of telomerase activity(GO:0051973) |
| 0.2 |
0.5 |
GO:0002268 |
follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.2 |
0.7 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.2 |
0.7 |
GO:0046599 |
regulation of centriole replication(GO:0046599) |
| 0.2 |
0.7 |
GO:0019087 |
transformation of host cell by virus(GO:0019087) |
| 0.2 |
3.6 |
GO:0009812 |
flavonoid metabolic process(GO:0009812) |
| 0.2 |
1.8 |
GO:1901985 |
positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.2 |
0.9 |
GO:0032933 |
response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 |
1.1 |
GO:0001306 |
age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.2 |
2.2 |
GO:0071285 |
cellular response to lithium ion(GO:0071285) |
| 0.2 |
1.3 |
GO:0006562 |
proline catabolic process(GO:0006562) |
| 0.2 |
0.7 |
GO:0010586 |
miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.2 |
1.3 |
GO:0009649 |
entrainment of circadian clock(GO:0009649) |
| 0.2 |
13.4 |
GO:0030183 |
B cell differentiation(GO:0030183) |
| 0.2 |
0.7 |
GO:0019242 |
methylglyoxal biosynthetic process(GO:0019242) |
| 0.2 |
0.6 |
GO:1903961 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.2 |
1.1 |
GO:0014805 |
smooth muscle adaptation(GO:0014805) |
| 0.2 |
14.2 |
GO:0001889 |
liver development(GO:0001889) |
| 0.2 |
1.3 |
GO:0043045 |
DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.2 |
0.2 |
GO:0071545 |
inositol phosphate catabolic process(GO:0071545) |
| 0.2 |
0.2 |
GO:1903426 |
regulation of nitric oxide biosynthetic process(GO:0045428) regulation of reactive oxygen species biosynthetic process(GO:1903426) |
| 0.2 |
0.6 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 |
0.4 |
GO:0000414 |
regulation of histone H3-K36 methylation(GO:0000414) |
| 0.2 |
0.4 |
GO:0002674 |
negative regulation of acute inflammatory response(GO:0002674) |
| 0.2 |
0.6 |
GO:0032074 |
negative regulation of nuclease activity(GO:0032074) |
| 0.2 |
1.0 |
GO:0021834 |
embryonic olfactory bulb interneuron precursor migration(GO:0021831) chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.2 |
0.8 |
GO:0097696 |
JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.2 |
0.4 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.2 |
0.2 |
GO:0001890 |
placenta development(GO:0001890) |
| 0.2 |
0.2 |
GO:0060279 |
regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.2 |
5.5 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.2 |
0.6 |
GO:0002637 |
regulation of immunoglobulin production(GO:0002637) positive regulation of immunoglobulin production(GO:0002639) |
| 0.2 |
1.0 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 |
2.1 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.2 |
0.4 |
GO:0033085 |
negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of thymocyte aggregation(GO:2000399) |
| 0.2 |
1.5 |
GO:0034755 |
iron ion transmembrane transport(GO:0034755) |
| 0.2 |
1.1 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.2 |
0.7 |
GO:0034086 |
maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.2 |
3.4 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.2 |
1.1 |
GO:0007063 |
regulation of sister chromatid cohesion(GO:0007063) |
| 0.2 |
1.3 |
GO:0033160 |
positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.2 |
0.7 |
GO:0045199 |
maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 |
0.5 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.2 |
0.5 |
GO:0010041 |
response to iron(III) ion(GO:0010041) positive regulation of histone phosphorylation(GO:0033129) response to DDT(GO:0046680) regulation of chromosome condensation(GO:0060623) cellular response to iron ion(GO:0071281) cellular response to iron(III) ion(GO:0071283) |
| 0.2 |
1.5 |
GO:0006787 |
porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.2 |
0.4 |
GO:0045780 |
positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.2 |
0.2 |
GO:0002562 |
somatic diversification of immune receptors via germline recombination within a single locus(GO:0002562) somatic cell DNA recombination(GO:0016444) |
| 0.2 |
0.2 |
GO:0032740 |
positive regulation of interleukin-17 production(GO:0032740) |
| 0.2 |
2.9 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.2 |
0.2 |
GO:0072053 |
renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.2 |
1.6 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.2 |
0.4 |
GO:2000309 |
tumor necrosis factor (ligand) superfamily member 11 production(GO:0072535) regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000307) positive regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000309) |
| 0.2 |
0.5 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 |
0.2 |
GO:0021898 |
commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.2 |
5.9 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.2 |
0.3 |
GO:0014040 |
regulation of Schwann cell differentiation(GO:0014038) positive regulation of Schwann cell differentiation(GO:0014040) |
| 0.2 |
0.7 |
GO:0018352 |
glutamate decarboxylation to succinate(GO:0006540) protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.2 |
1.6 |
GO:0018126 |
protein hydroxylation(GO:0018126) |
| 0.2 |
5.0 |
GO:0030593 |
neutrophil chemotaxis(GO:0030593) neutrophil migration(GO:1990266) |
| 0.2 |
0.2 |
GO:2000273 |
positive regulation of receptor activity(GO:2000273) |
| 0.2 |
1.7 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.2 |
0.9 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.2 |
0.7 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.2 |
0.7 |
GO:0060046 |
regulation of acrosome reaction(GO:0060046) |
| 0.2 |
0.7 |
GO:0006408 |
snRNA export from nucleus(GO:0006408) snRNA transport(GO:0051030) |
| 0.2 |
0.5 |
GO:0015854 |
guanine transport(GO:0015854) hypoxanthine transport(GO:0035344) thymine transport(GO:0035364) |
| 0.2 |
0.2 |
GO:0019369 |
arachidonic acid metabolic process(GO:0019369) |
| 0.2 |
0.5 |
GO:0060674 |
placenta blood vessel development(GO:0060674) |
| 0.2 |
0.5 |
GO:0003351 |
epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 |
1.0 |
GO:0015871 |
choline transport(GO:0015871) |
| 0.2 |
3.6 |
GO:0002504 |
antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.2 |
0.8 |
GO:0019852 |
L-ascorbic acid metabolic process(GO:0019852) |
| 0.2 |
0.5 |
GO:0060397 |
JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 |
0.3 |
GO:0097237 |
response to cobalt ion(GO:0032025) cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.2 |
0.5 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.2 |
0.5 |
GO:0007500 |
mesodermal cell fate determination(GO:0007500) |
| 0.2 |
0.8 |
GO:0035455 |
response to interferon-alpha(GO:0035455) |
| 0.2 |
3.6 |
GO:0090630 |
activation of GTPase activity(GO:0090630) |
| 0.2 |
0.3 |
GO:0015959 |
diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.2 |
0.2 |
GO:0032203 |
telomere formation via telomerase(GO:0032203) |
| 0.2 |
0.9 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.2 |
0.5 |
GO:0071679 |
facial nucleus development(GO:0021754) commissural neuron axon guidance(GO:0071679) |
| 0.2 |
0.5 |
GO:1901419 |
regulation of response to alcohol(GO:1901419) |
| 0.2 |
0.6 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 |
5.7 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.2 |
0.2 |
GO:0021854 |
hypothalamus development(GO:0021854) |
| 0.2 |
4.5 |
GO:0031100 |
organ regeneration(GO:0031100) |
| 0.2 |
0.3 |
GO:0032200 |
telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.2 |
0.9 |
GO:0048296 |
isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 |
0.3 |
GO:0010159 |
specification of organ position(GO:0010159) |
| 0.1 |
0.6 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 |
0.7 |
GO:0051852 |
disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.1 |
0.7 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 |
0.6 |
GO:0043116 |
negative regulation of vascular permeability(GO:0043116) |
| 0.1 |
0.4 |
GO:1901142 |
insulin processing(GO:0030070) insulin metabolic process(GO:1901142) |
| 0.1 |
2.3 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.1 |
0.6 |
GO:0042637 |
catagen(GO:0042637) |
| 0.1 |
0.7 |
GO:0003416 |
endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.1 |
2.2 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.1 |
0.4 |
GO:0031998 |
regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 |
1.7 |
GO:0006260 |
DNA replication(GO:0006260) |
| 0.1 |
1.3 |
GO:0030147 |
obsolete natriuresis(GO:0030147) |
| 0.1 |
0.4 |
GO:0048537 |
mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.1 |
0.4 |
GO:0071168 |
protein localization to chromatin(GO:0071168) |
| 0.1 |
0.4 |
GO:0070309 |
lens fiber cell morphogenesis(GO:0070309) |
| 0.1 |
0.1 |
GO:0046851 |
negative regulation of tissue remodeling(GO:0034104) negative regulation of bone resorption(GO:0045779) negative regulation of bone remodeling(GO:0046851) |
| 0.1 |
0.4 |
GO:0035330 |
regulation of hippo signaling(GO:0035330) |
| 0.1 |
0.4 |
GO:0045091 |
regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.1 |
0.1 |
GO:0051531 |
NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.1 |
0.4 |
GO:0006167 |
AMP biosynthetic process(GO:0006167) |
| 0.1 |
0.4 |
GO:0071848 |
regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) TNFSF11-mediated signaling pathway(GO:0071847) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.1 |
1.1 |
GO:0046498 |
S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.1 |
0.3 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 |
1.5 |
GO:0042738 |
exogenous drug catabolic process(GO:0042738) |
| 0.1 |
12.6 |
GO:0007605 |
sensory perception of sound(GO:0007605) |
| 0.1 |
0.3 |
GO:0010572 |
positive regulation of platelet activation(GO:0010572) |
| 0.1 |
1.1 |
GO:0031954 |
positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 |
0.4 |
GO:0032223 |
negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 |
0.9 |
GO:0016075 |
rRNA catabolic process(GO:0016075) |
| 0.1 |
0.1 |
GO:0051928 |
positive regulation of calcium ion transport(GO:0051928) |
| 0.1 |
0.5 |
GO:0060192 |
negative regulation of lipoprotein lipase activity(GO:0051005) negative regulation of lipase activity(GO:0060192) |
| 0.1 |
0.4 |
GO:0018916 |
nitrobenzene metabolic process(GO:0018916) |
| 0.1 |
0.7 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.1 |
1.0 |
GO:0042992 |
negative regulation of transcription factor import into nucleus(GO:0042992) |
| 0.1 |
1.2 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.1 |
0.3 |
GO:2000051 |
negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 |
0.3 |
GO:0010765 |
positive regulation of sodium ion transport(GO:0010765) |
| 0.1 |
1.2 |
GO:0090080 |
positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 |
0.4 |
GO:0006536 |
glutamate metabolic process(GO:0006536) |
| 0.1 |
0.6 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 |
0.8 |
GO:0046683 |
response to organophosphorus(GO:0046683) |
| 0.1 |
0.5 |
GO:0000212 |
meiotic spindle organization(GO:0000212) |
| 0.1 |
0.7 |
GO:0034616 |
response to laminar fluid shear stress(GO:0034616) |
| 0.1 |
0.7 |
GO:0045072 |
regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 |
0.7 |
GO:0010866 |
regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.1 |
0.6 |
GO:0045767 |
obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.1 |
4.3 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.1 |
0.2 |
GO:0044240 |
multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 |
0.6 |
GO:0010957 |
negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 |
0.4 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 |
0.7 |
GO:0001840 |
neural plate morphogenesis(GO:0001839) neural plate development(GO:0001840) |
| 0.1 |
0.6 |
GO:0051323 |
metaphase(GO:0051323) |
| 0.1 |
0.7 |
GO:0071496 |
cellular response to external stimulus(GO:0071496) |
| 0.1 |
0.2 |
GO:0030099 |
myeloid cell differentiation(GO:0030099) |
| 0.1 |
0.8 |
GO:0042246 |
tissue regeneration(GO:0042246) |
| 0.1 |
1.1 |
GO:0008089 |
anterograde axonal transport(GO:0008089) |
| 0.1 |
2.8 |
GO:0060334 |
regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 |
0.7 |
GO:0043615 |
astrocyte cell migration(GO:0043615) |
| 0.1 |
1.5 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.1 |
2.3 |
GO:0046676 |
negative regulation of insulin secretion(GO:0046676) |
| 0.1 |
0.5 |
GO:0002544 |
chronic inflammatory response(GO:0002544) |
| 0.1 |
0.1 |
GO:0032609 |
interferon-gamma production(GO:0032609) regulation of interferon-gamma production(GO:0032649) |
| 0.1 |
2.9 |
GO:0033209 |
tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 |
0.7 |
GO:0048302 |
isotype switching to IgG isotypes(GO:0048291) regulation of isotype switching to IgG isotypes(GO:0048302) |
| 0.1 |
0.9 |
GO:0007080 |
mitotic metaphase plate congression(GO:0007080) |
| 0.1 |
5.9 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.1 |
0.5 |
GO:0060649 |
nipple development(GO:0060618) mammary gland bud elongation(GO:0060649) nipple morphogenesis(GO:0060658) nipple sheath formation(GO:0060659) |
| 0.1 |
0.3 |
GO:0042095 |
interferon-gamma biosynthetic process(GO:0042095) |
| 0.1 |
0.7 |
GO:0050684 |
regulation of mRNA processing(GO:0050684) regulation of mRNA metabolic process(GO:1903311) |
| 0.1 |
0.2 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.1 |
1.5 |
GO:0045730 |
respiratory burst(GO:0045730) |
| 0.1 |
2.1 |
GO:0002026 |
regulation of the force of heart contraction(GO:0002026) |
| 0.1 |
0.1 |
GO:0045835 |
negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 |
0.7 |
GO:0046835 |
carbohydrate phosphorylation(GO:0046835) |
| 0.1 |
0.2 |
GO:0009648 |
photoperiodism(GO:0009648) |
| 0.1 |
0.2 |
GO:0046967 |
cytosol to ER transport(GO:0046967) |
| 0.1 |
1.0 |
GO:1900087 |
traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.1 |
0.3 |
GO:0007518 |
myoblast fate determination(GO:0007518) |
| 0.1 |
0.3 |
GO:0008298 |
intracellular mRNA localization(GO:0008298) |
| 0.1 |
1.4 |
GO:0010039 |
response to iron ion(GO:0010039) |
| 0.1 |
0.1 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.1 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 |
0.2 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 |
1.0 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.1 |
0.4 |
GO:0016074 |
snoRNA metabolic process(GO:0016074) |
| 0.1 |
1.8 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.1 |
0.8 |
GO:0042832 |
defense response to protozoan(GO:0042832) |
| 0.1 |
0.5 |
GO:0031639 |
plasminogen activation(GO:0031639) |
| 0.1 |
0.6 |
GO:0046653 |
tetrahydrofolate metabolic process(GO:0046653) |
| 0.1 |
1.2 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.1 |
1.4 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.1 |
1.6 |
GO:0046365 |
monosaccharide catabolic process(GO:0046365) |
| 0.1 |
0.6 |
GO:0032814 |
regulation of natural killer cell activation(GO:0032814) |
| 0.1 |
0.3 |
GO:0048478 |
replication fork protection(GO:0048478) |
| 0.1 |
0.9 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.1 |
1.8 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 |
0.1 |
GO:0016554 |
cytidine to uridine editing(GO:0016554) |
| 0.1 |
0.4 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 |
0.7 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.1 |
0.8 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.1 |
0.8 |
GO:0006600 |
creatine metabolic process(GO:0006600) |
| 0.1 |
0.8 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.1 |
0.3 |
GO:0016476 |
regulation of embryonic cell shape(GO:0016476) |
| 0.1 |
0.6 |
GO:0051917 |
regulation of fibrinolysis(GO:0051917) negative regulation of fibrinolysis(GO:0051918) |
| 0.1 |
0.4 |
GO:0045338 |
farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 |
0.9 |
GO:0016540 |
protein autoprocessing(GO:0016540) |
| 0.1 |
0.1 |
GO:0007584 |
response to nutrient(GO:0007584) |
| 0.1 |
0.2 |
GO:0021549 |
cerebellum development(GO:0021549) |
| 0.1 |
2.1 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.1 |
0.2 |
GO:0051971 |
positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.1 |
0.3 |
GO:0035608 |
protein deglutamylation(GO:0035608) |
| 0.1 |
0.4 |
GO:0032604 |
granulocyte macrophage colony-stimulating factor production(GO:0032604) regulation of granulocyte macrophage colony-stimulating factor production(GO:0032645) positive regulation of granulocyte macrophage colony-stimulating factor production(GO:0032725) |
| 0.1 |
0.8 |
GO:0006670 |
sphingosine metabolic process(GO:0006670) diol metabolic process(GO:0034311) |
| 0.1 |
0.3 |
GO:0032927 |
positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 |
0.2 |
GO:0042534 |
tumor necrosis factor biosynthetic process(GO:0042533) regulation of tumor necrosis factor biosynthetic process(GO:0042534) |
| 0.1 |
0.1 |
GO:0051582 |
positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.1 |
0.2 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 |
0.7 |
GO:0000380 |
alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 |
1.5 |
GO:0006582 |
melanin metabolic process(GO:0006582) |
| 0.1 |
0.4 |
GO:0060430 |
globus pallidus development(GO:0021759) menarche(GO:0042696) lung saccule development(GO:0060430) Clara cell differentiation(GO:0060486) Type II pneumocyte differentiation(GO:0060510) |
| 0.1 |
0.1 |
GO:0043628 |
ncRNA 3'-end processing(GO:0043628) |
| 0.1 |
0.7 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.1 |
0.4 |
GO:0019348 |
polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.1 |
0.3 |
GO:0045953 |
negative regulation of natural killer cell mediated immunity(GO:0002716) negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.1 |
0.6 |
GO:0001782 |
B cell homeostasis(GO:0001782) |
| 0.1 |
0.3 |
GO:1901224 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 |
0.2 |
GO:0060398 |
regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.1 |
0.4 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.1 |
0.1 |
GO:0006505 |
GPI anchor metabolic process(GO:0006505) |
| 0.1 |
0.8 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.1 |
0.1 |
GO:0033599 |
regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.1 |
0.3 |
GO:0000050 |
urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.1 |
0.4 |
GO:0033261 |
obsolete regulation of S phase(GO:0033261) |
| 0.1 |
3.1 |
GO:0043124 |
negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 |
0.3 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.1 |
0.4 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.1 |
1.2 |
GO:0051568 |
histone H3-K4 methylation(GO:0051568) |
| 0.1 |
0.3 |
GO:0010759 |
regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.1 |
1.1 |
GO:0070542 |
response to fatty acid(GO:0070542) |
| 0.1 |
1.0 |
GO:0010259 |
multicellular organism aging(GO:0010259) |
| 0.1 |
1.1 |
GO:0019221 |
cytokine-mediated signaling pathway(GO:0019221) |
| 0.1 |
2.2 |
GO:0051092 |
positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 |
1.1 |
GO:0051923 |
sulfation(GO:0051923) |
| 0.1 |
0.5 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 |
0.3 |
GO:0015809 |
arginine transport(GO:0015809) |
| 0.1 |
0.1 |
GO:0016447 |
somatic recombination of immunoglobulin gene segments(GO:0016447) |
| 0.1 |
0.6 |
GO:0050812 |
acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 |
1.9 |
GO:0006271 |
DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 |
0.4 |
GO:0006105 |
succinate metabolic process(GO:0006105) |
| 0.1 |
0.8 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.1 |
0.2 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 |
0.7 |
GO:0045197 |
establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 |
0.2 |
GO:0070254 |
mucus secretion(GO:0070254) |
| 0.1 |
0.2 |
GO:0045618 |
positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 |
2.5 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.1 |
0.2 |
GO:0048845 |
venous blood vessel morphogenesis(GO:0048845) |
| 0.1 |
0.6 |
GO:0005977 |
polysaccharide metabolic process(GO:0005976) glycogen metabolic process(GO:0005977) cellular polysaccharide metabolic process(GO:0044264) |
| 0.1 |
0.5 |
GO:0043316 |
natural killer cell activation involved in immune response(GO:0002323) cytotoxic T cell degranulation(GO:0043316) natural killer cell degranulation(GO:0043320) |
| 0.1 |
0.2 |
GO:0042102 |
positive regulation of T cell proliferation(GO:0042102) |
| 0.1 |
0.1 |
GO:1903514 |
regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.1 |
0.6 |
GO:0014046 |
dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 |
0.4 |
GO:0015803 |
aromatic amino acid transport(GO:0015801) branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) tryptophan transport(GO:0015827) leucine import(GO:0060356) |
| 0.1 |
0.5 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.1 |
0.2 |
GO:0070849 |
response to epidermal growth factor(GO:0070849) |
| 0.1 |
0.9 |
GO:0030010 |
establishment of cell polarity(GO:0030010) |
| 0.1 |
0.2 |
GO:0045759 |
negative regulation of action potential(GO:0045759) |
| 0.1 |
3.6 |
GO:0016337 |
single organismal cell-cell adhesion(GO:0016337) |
| 0.1 |
0.4 |
GO:0006896 |
Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.1 |
1.5 |
GO:0009067 |
aspartate family amino acid biosynthetic process(GO:0009067) |
| 0.1 |
0.2 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.1 |
0.6 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 |
1.3 |
GO:0042255 |
ribosome assembly(GO:0042255) |
| 0.1 |
0.3 |
GO:0051958 |
methotrexate transport(GO:0051958) |
| 0.1 |
0.2 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 |
1.9 |
GO:0046580 |
negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 |
1.6 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 |
0.2 |
GO:0034447 |
very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.1 |
4.8 |
GO:0031647 |
regulation of protein stability(GO:0031647) |
| 0.1 |
0.2 |
GO:0060042 |
retina morphogenesis in camera-type eye(GO:0060042) |
| 0.1 |
0.2 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
0.6 |
GO:0045008 |
depyrimidination(GO:0045008) |
| 0.1 |
0.4 |
GO:0070537 |
histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 |
1.0 |
GO:0050930 |
induction of positive chemotaxis(GO:0050930) |
| 0.1 |
0.4 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 |
0.1 |
GO:0007398 |
ectoderm development(GO:0007398) |
| 0.1 |
0.4 |
GO:0016064 |
immunoglobulin mediated immune response(GO:0016064) |
| 0.1 |
0.1 |
GO:0052556 |
pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) positive regulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052555) positive regulation by symbiont of host immune response(GO:0052556) |
| 0.1 |
0.2 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.1 |
4.8 |
GO:0006821 |
chloride transport(GO:0006821) |
| 0.1 |
1.4 |
GO:0006342 |
chromatin silencing(GO:0006342) |
| 0.1 |
0.3 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.1 |
0.4 |
GO:0048385 |
regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.1 |
0.3 |
GO:0046642 |
negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 |
0.4 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 |
9.6 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 |
3.4 |
GO:0042113 |
B cell activation(GO:0042113) |
| 0.1 |
1.9 |
GO:0005980 |
glycogen catabolic process(GO:0005980) |
| 0.1 |
0.3 |
GO:0001539 |
cilium or flagellum-dependent cell motility(GO:0001539) |
| 0.1 |
0.8 |
GO:0030219 |
megakaryocyte differentiation(GO:0030219) |
| 0.1 |
0.1 |
GO:0003077 |
obsolete negative regulation of diuresis(GO:0003077) |
| 0.1 |
0.5 |
GO:0060314 |
regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 |
0.1 |
GO:0006782 |
protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.1 |
0.3 |
GO:0030037 |
actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.1 |
1.3 |
GO:0006386 |
transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.1 |
0.5 |
GO:0051293 |
establishment of mitotic spindle localization(GO:0040001) establishment of spindle localization(GO:0051293) spindle localization(GO:0051653) |
| 0.1 |
1.3 |
GO:0007257 |
activation of JUN kinase activity(GO:0007257) |
| 0.1 |
0.2 |
GO:0060487 |
lung cell differentiation(GO:0060479) lung epithelial cell differentiation(GO:0060487) |
| 0.1 |
0.8 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
0.1 |
GO:0060689 |
cell differentiation involved in salivary gland development(GO:0060689) |
| 0.1 |
0.1 |
GO:0032096 |
negative regulation of response to food(GO:0032096) |
| 0.1 |
0.3 |
GO:0071354 |
cellular response to interleukin-6(GO:0071354) |
| 0.1 |
0.5 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 |
1.9 |
GO:0030216 |
keratinocyte differentiation(GO:0030216) |
| 0.1 |
0.2 |
GO:0071371 |
response to follicle-stimulating hormone(GO:0032354) cellular response to gonadotropin stimulus(GO:0071371) cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 |
0.2 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 |
0.5 |
GO:0050709 |
negative regulation of protein secretion(GO:0050709) |
| 0.1 |
0.3 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 |
0.3 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 |
0.1 |
GO:0060484 |
lung-associated mesenchyme development(GO:0060484) |
| 0.1 |
2.1 |
GO:0001892 |
embryonic placenta development(GO:0001892) |
| 0.1 |
0.2 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 |
0.1 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.1 |
0.2 |
GO:0055003 |
cardiac myofibril assembly(GO:0055003) |
| 0.1 |
0.8 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 |
0.3 |
GO:0018106 |
peptidyl-histidine phosphorylation(GO:0018106) |
| 0.1 |
1.0 |
GO:0032098 |
regulation of appetite(GO:0032098) |
| 0.1 |
2.2 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 |
0.3 |
GO:0006998 |
nuclear envelope organization(GO:0006998) |
| 0.1 |
0.5 |
GO:0032651 |
regulation of interleukin-1 beta production(GO:0032651) |
| 0.1 |
0.7 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.1 |
0.4 |
GO:0006560 |
proline metabolic process(GO:0006560) |
| 0.1 |
0.9 |
GO:0042593 |
carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.1 |
0.3 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 |
0.1 |
GO:0045815 |
positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.1 |
1.5 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.1 |
3.4 |
GO:0072164 |
ureteric bud development(GO:0001657) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.1 |
0.4 |
GO:0031444 |
slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 |
4.2 |
GO:0071357 |
response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 |
0.6 |
GO:1901663 |
quinone biosynthetic process(GO:1901663) |
| 0.1 |
0.2 |
GO:0061525 |
hindgut morphogenesis(GO:0007442) hindgut development(GO:0061525) |
| 0.1 |
0.2 |
GO:0051307 |
resolution of meiotic recombination intermediates(GO:0000712) meiotic chromosome separation(GO:0051307) |
| 0.1 |
0.2 |
GO:0000114 |
obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.1 |
0.2 |
GO:1904396 |
regulation of synaptic growth at neuromuscular junction(GO:0008582) regulation of neuromuscular junction development(GO:1904396) |
| 0.1 |
0.6 |
GO:2000117 |
negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.1 |
1.8 |
GO:0050909 |
sensory perception of taste(GO:0050909) |
| 0.1 |
0.1 |
GO:0008212 |
mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.1 |
0.7 |
GO:0048705 |
skeletal system morphogenesis(GO:0048705) |
| 0.1 |
0.2 |
GO:0021877 |
forebrain neuron fate commitment(GO:0021877) |
| 0.1 |
0.2 |
GO:0043418 |
cysteine biosynthetic process from serine(GO:0006535) homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) homocysteine catabolic process(GO:0043418) hydrogen sulfide metabolic process(GO:0070813) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 |
0.2 |
GO:0001938 |
positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.1 |
0.6 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.1 |
0.2 |
GO:0045948 |
positive regulation of translational initiation(GO:0045948) |
| 0.1 |
0.1 |
GO:0031018 |
endocrine pancreas development(GO:0031018) |
| 0.1 |
0.1 |
GO:0001880 |
Mullerian duct regression(GO:0001880) anatomical structure regression(GO:0060033) |
| 0.1 |
0.7 |
GO:0000060 |
protein import into nucleus, translocation(GO:0000060) |
| 0.1 |
0.6 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 |
0.2 |
GO:0009607 |
response to biotic stimulus(GO:0009607) |
| 0.1 |
0.2 |
GO:0060405 |
regulation of penile erection(GO:0060405) |
| 0.1 |
0.1 |
GO:0010891 |
negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 |
0.2 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 |
0.4 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 |
0.1 |
GO:0045351 |
type I interferon biosynthetic process(GO:0045351) |
| 0.1 |
0.2 |
GO:0034219 |
carbohydrate transmembrane transport(GO:0034219) |
| 0.1 |
0.4 |
GO:0048712 |
negative regulation of astrocyte differentiation(GO:0048712) |
| 0.1 |
0.2 |
GO:0030238 |
male sex determination(GO:0030238) |
| 0.1 |
1.0 |
GO:0007405 |
neuroblast proliferation(GO:0007405) |
| 0.1 |
0.1 |
GO:0001507 |
acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.1 |
0.4 |
GO:0045745 |
positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 |
0.4 |
GO:0032464 |
positive regulation of protein oligomerization(GO:0032461) positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 |
0.3 |
GO:0042993 |
positive regulation of NF-kappaB import into nucleus(GO:0042346) positive regulation of transcription factor import into nucleus(GO:0042993) |
| 0.1 |
0.2 |
GO:0000394 |
RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 |
0.6 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.1 |
0.2 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 |
0.1 |
GO:0061756 |
leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.1 |
1.7 |
GO:0007602 |
phototransduction(GO:0007602) |
| 0.1 |
0.1 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.1 |
0.4 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.1 |
0.2 |
GO:0032494 |
response to peptidoglycan(GO:0032494) |
| 0.1 |
0.3 |
GO:0040023 |
establishment of nucleus localization(GO:0040023) |
| 0.1 |
0.2 |
GO:1903115 |
regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.1 |
0.5 |
GO:0035036 |
binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 |
0.2 |
GO:0007351 |
blastoderm segmentation(GO:0007350) tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 |
0.7 |
GO:0006400 |
tRNA modification(GO:0006400) |
| 0.0 |
0.1 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) |
| 0.0 |
0.1 |
GO:0032229 |
negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 |
0.1 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.0 |
0.1 |
GO:0051547 |
regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 |
0.2 |
GO:2000171 |
negative regulation of dendrite development(GO:2000171) |
| 0.0 |
0.1 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 |
0.2 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.0 |
0.0 |
GO:0015742 |
alpha-ketoglutarate transport(GO:0015742) |
| 0.0 |
0.9 |
GO:0006596 |
polyamine biosynthetic process(GO:0006596) |
| 0.0 |
0.9 |
GO:0030851 |
granulocyte differentiation(GO:0030851) |
| 0.0 |
0.1 |
GO:0050798 |
activated T cell proliferation(GO:0050798) |
| 0.0 |
0.3 |
GO:0018027 |
peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 |
0.1 |
GO:0032808 |
lacrimal gland development(GO:0032808) Harderian gland development(GO:0070384) |
| 0.0 |
0.1 |
GO:0021940 |
regulation of cerebellar granule cell precursor proliferation(GO:0021936) cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.0 |
0.1 |
GO:0030997 |
regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 |
0.1 |
GO:0032768 |
regulation of monooxygenase activity(GO:0032768) |
| 0.0 |
0.9 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.5 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 |
0.1 |
GO:0043501 |
skeletal muscle adaptation(GO:0043501) |
| 0.0 |
0.0 |
GO:0051602 |
response to electrical stimulus(GO:0051602) |
| 0.0 |
0.1 |
GO:0021983 |
pituitary gland development(GO:0021983) |
| 0.0 |
0.3 |
GO:0030308 |
negative regulation of cell growth(GO:0030308) negative regulation of growth(GO:0045926) |
| 0.0 |
0.1 |
GO:0001702 |
gastrulation with mouth forming second(GO:0001702) |
| 0.0 |
0.3 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.6 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
1.2 |
GO:0030262 |
apoptotic nuclear changes(GO:0030262) |
| 0.0 |
0.1 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.0 |
0.1 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.0 |
0.1 |
GO:0034442 |
plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.0 |
0.2 |
GO:1901976 |
regulation of spindle checkpoint(GO:0090231) regulation of cell cycle checkpoint(GO:1901976) |
| 0.0 |
0.4 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.1 |
GO:0045898 |
regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 |
2.1 |
GO:0051084 |
'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 |
0.1 |
GO:0014888 |
striated muscle adaptation(GO:0014888) |
| 0.0 |
0.2 |
GO:0002260 |
lymphocyte homeostasis(GO:0002260) |
| 0.0 |
0.6 |
GO:0010458 |
exit from mitosis(GO:0010458) |
| 0.0 |
1.6 |
GO:0006094 |
gluconeogenesis(GO:0006094) |
| 0.0 |
0.5 |
GO:0006310 |
DNA recombination(GO:0006310) |
| 0.0 |
0.1 |
GO:0071377 |
cellular response to glucagon stimulus(GO:0071377) |
| 0.0 |
0.1 |
GO:0000966 |
RNA 5'-end processing(GO:0000966) |
| 0.0 |
0.3 |
GO:0006084 |
acetyl-CoA metabolic process(GO:0006084) |
| 0.0 |
0.6 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.0 |
0.0 |
GO:0036260 |
7-methylguanosine mRNA capping(GO:0006370) 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 |
0.7 |
GO:0050830 |
defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 |
2.1 |
GO:0008654 |
phospholipid biosynthetic process(GO:0008654) |
| 0.0 |
0.3 |
GO:0032916 |
transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 |
0.0 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 |
0.5 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.0 |
5.7 |
GO:0043087 |
regulation of GTPase activity(GO:0043087) |
| 0.0 |
0.5 |
GO:0019752 |
carboxylic acid metabolic process(GO:0019752) |
| 0.0 |
0.1 |
GO:0030320 |
cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) |
| 0.0 |
0.2 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.0 |
0.2 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.3 |
GO:0009451 |
RNA modification(GO:0009451) |
| 0.0 |
0.2 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.8 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
0.1 |
GO:0042363 |
vitamin catabolic process(GO:0009111) fat-soluble vitamin catabolic process(GO:0042363) |
| 0.0 |
1.5 |
GO:0042742 |
defense response to bacterium(GO:0042742) |
| 0.0 |
1.2 |
GO:0007051 |
spindle organization(GO:0007051) |
| 0.0 |
0.3 |
GO:0006072 |
glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 |
1.1 |
GO:0000725 |
double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 |
0.0 |
GO:0060746 |
maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 |
0.4 |
GO:0051322 |
anaphase(GO:0051322) |
| 0.0 |
0.3 |
GO:0060770 |
regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 |
0.8 |
GO:0031055 |
DNA replication-independent nucleosome assembly(GO:0006336) chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) DNA replication-independent nucleosome organization(GO:0034724) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 |
0.1 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 |
2.1 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.1 |
GO:0046689 |
response to mercury ion(GO:0046689) |
| 0.0 |
0.2 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 |
0.4 |
GO:0048546 |
digestive tract morphogenesis(GO:0048546) |
| 0.0 |
0.4 |
GO:0008635 |
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 |
3.0 |
GO:0006364 |
rRNA processing(GO:0006364) rRNA metabolic process(GO:0016072) |
| 0.0 |
0.3 |
GO:0042596 |
fear response(GO:0042596) |
| 0.0 |
0.2 |
GO:0050858 |
negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.1 |
GO:0060152 |
peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 |
0.2 |
GO:0021978 |
telencephalon regionalization(GO:0021978) |
| 0.0 |
1.1 |
GO:0032436 |
positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 |
0.3 |
GO:0019987 |
obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 |
0.8 |
GO:0006691 |
leukotriene metabolic process(GO:0006691) |
| 0.0 |
1.0 |
GO:0007193 |
adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 |
0.3 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.2 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.0 |
0.1 |
GO:0031000 |
response to caffeine(GO:0031000) |
| 0.0 |
0.2 |
GO:0061448 |
connective tissue development(GO:0061448) |
| 0.0 |
0.3 |
GO:0006308 |
DNA catabolic process(GO:0006308) |
| 0.0 |
0.2 |
GO:0048536 |
spleen development(GO:0048536) |
| 0.0 |
0.2 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
0.1 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 |
0.3 |
GO:0032411 |
positive regulation of transporter activity(GO:0032411) |
| 0.0 |
0.1 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 |
0.1 |
GO:0046015 |
carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 |
0.3 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.3 |
GO:0051354 |
negative regulation of oxidoreductase activity(GO:0051354) |
| 0.0 |
0.1 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.3 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.0 |
0.1 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) |
| 0.0 |
1.8 |
GO:0006338 |
chromatin remodeling(GO:0006338) |
| 0.0 |
0.1 |
GO:0045872 |
positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 |
0.1 |
GO:0000076 |
DNA replication checkpoint(GO:0000076) |
| 0.0 |
1.2 |
GO:0006383 |
transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 |
0.4 |
GO:0051703 |
social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 |
0.1 |
GO:0016926 |
protein desumoylation(GO:0016926) |
| 0.0 |
0.3 |
GO:0006469 |
negative regulation of protein kinase activity(GO:0006469) |
| 0.0 |
0.2 |
GO:0033962 |
cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 |
0.1 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.0 |
2.7 |
GO:0050852 |
T cell receptor signaling pathway(GO:0050852) |
| 0.0 |
1.1 |
GO:0019079 |
viral genome replication(GO:0019079) |
| 0.0 |
0.1 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 |
0.4 |
GO:0046834 |
lipid phosphorylation(GO:0046834) |
| 0.0 |
0.1 |
GO:0046463 |
triglyceride biosynthetic process(GO:0019432) neutral lipid biosynthetic process(GO:0046460) acylglycerol biosynthetic process(GO:0046463) |
| 0.0 |
0.2 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 |
0.1 |
GO:0016101 |
retinoid metabolic process(GO:0001523) diterpenoid metabolic process(GO:0016101) |
| 0.0 |
0.1 |
GO:0031646 |
positive regulation of neurological system process(GO:0031646) |
| 0.0 |
0.3 |
GO:0043507 |
positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 |
0.1 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.0 |
0.1 |
GO:0014823 |
response to activity(GO:0014823) |
| 0.0 |
0.3 |
GO:0007062 |
sister chromatid cohesion(GO:0007062) |
| 0.0 |
0.1 |
GO:0045814 |
negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 |
0.1 |
GO:0006884 |
cell volume homeostasis(GO:0006884) |
| 0.0 |
0.3 |
GO:0030101 |
natural killer cell activation(GO:0030101) |
| 0.0 |
0.1 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.7 |
GO:0016254 |
preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 |
0.0 |
GO:0032611 |
interleukin-1 beta production(GO:0032611) |
| 0.0 |
0.1 |
GO:0043278 |
response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 |
0.2 |
GO:0042416 |
dopamine biosynthetic process(GO:0042416) |
| 0.0 |
0.2 |
GO:1901998 |
tetracycline transport(GO:0015904) antibiotic transport(GO:0042891) toxin transport(GO:1901998) |
| 0.0 |
0.1 |
GO:1903078 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 |
0.1 |
GO:0048284 |
organelle fusion(GO:0048284) organelle membrane fusion(GO:0090174) |
| 0.0 |
0.2 |
GO:0051881 |
regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 |
0.1 |
GO:0048935 |
peripheral nervous system neuron development(GO:0048935) |
| 0.0 |
0.1 |
GO:0007549 |
dosage compensation(GO:0007549) |
| 0.0 |
2.1 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.3 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.0 |
0.1 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.0 |
0.3 |
GO:0000045 |
autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 |
0.1 |
GO:0060346 |
bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 |
0.2 |
GO:0007628 |
adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 |
0.7 |
GO:0042254 |
ribosome biogenesis(GO:0042254) |
| 0.0 |
0.1 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 |
0.5 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.0 |
0.2 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 |
0.0 |
GO:0001707 |
mesoderm formation(GO:0001707) mesoderm morphogenesis(GO:0048332) |
| 0.0 |
0.1 |
GO:0046685 |
response to arsenic-containing substance(GO:0046685) |
| 0.0 |
0.3 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.0 |
4.4 |
GO:0006813 |
potassium ion transport(GO:0006813) |
| 0.0 |
1.0 |
GO:0016579 |
protein deubiquitination(GO:0016579) |
| 0.0 |
0.8 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 |
1.8 |
GO:0000236 |
mitotic prometaphase(GO:0000236) |
| 0.0 |
0.1 |
GO:0030878 |
thyroid gland development(GO:0030878) |
| 0.0 |
0.2 |
GO:0007162 |
negative regulation of cell adhesion(GO:0007162) |
| 0.0 |
0.2 |
GO:0009128 |
AMP catabolic process(GO:0006196) purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 |
0.1 |
GO:0045086 |
positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 |
0.1 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.0 |
0.1 |
GO:0090274 |
somatostatin secretion(GO:0070253) regulation of somatostatin secretion(GO:0090273) positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 |
1.4 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.0 |
0.0 |
GO:0021520 |
spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 |
0.7 |
GO:0007422 |
peripheral nervous system development(GO:0007422) |
| 0.0 |
0.1 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.0 |
1.6 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.0 |
0.2 |
GO:0006474 |
N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 |
0.4 |
GO:0007346 |
regulation of mitotic cell cycle(GO:0007346) |
| 0.0 |
0.4 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.0 |
0.4 |
GO:0030317 |
sperm motility(GO:0030317) |
| 0.0 |
0.1 |
GO:0035413 |
positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 |
0.2 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.0 |
0.1 |
GO:0010595 |
positive regulation of endothelial cell migration(GO:0010595) |
| 0.0 |
0.1 |
GO:0022605 |
ovarian cumulus expansion(GO:0001550) oogenesis stage(GO:0022605) fused antrum stage(GO:0048165) |
| 0.0 |
0.1 |
GO:0048670 |
regulation of collateral sprouting(GO:0048670) |
| 0.0 |
0.2 |
GO:0031268 |
pseudopodium organization(GO:0031268) |
| 0.0 |
0.2 |
GO:0034383 |
low-density lipoprotein particle clearance(GO:0034383) |
| 0.0 |
0.1 |
GO:0016265 |
obsolete death(GO:0016265) |
| 0.0 |
0.1 |
GO:0046368 |
GDP-L-fucose biosynthetic process(GO:0042350) 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) GDP-L-fucose metabolic process(GO:0046368) |
| 0.0 |
0.2 |
GO:0070189 |
kynurenine metabolic process(GO:0070189) |
| 0.0 |
0.2 |
GO:0015939 |
pantothenate metabolic process(GO:0015939) |
| 0.0 |
0.1 |
GO:0015705 |
iodide transport(GO:0015705) |
| 0.0 |
0.1 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.1 |
GO:0000239 |
pachytene(GO:0000239) |
| 0.0 |
0.1 |
GO:0030299 |
intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 |
0.1 |
GO:0043525 |
positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 |
0.2 |
GO:0015695 |
organic cation transport(GO:0015695) |
| 0.0 |
0.1 |
GO:0006570 |
tyrosine metabolic process(GO:0006570) |
| 0.0 |
0.1 |
GO:0005996 |
monosaccharide metabolic process(GO:0005996) |
| 0.0 |
0.3 |
GO:0006406 |
mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 |
0.4 |
GO:0042771 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 |
0.2 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
1.8 |
GO:0006401 |
RNA catabolic process(GO:0006401) |
| 0.0 |
0.2 |
GO:0021675 |
nerve development(GO:0021675) |
| 0.0 |
0.2 |
GO:0001961 |
positive regulation of cytokine-mediated signaling pathway(GO:0001961) |
| 0.0 |
0.8 |
GO:0008033 |
tRNA processing(GO:0008033) |
| 0.0 |
3.2 |
GO:0006470 |
protein dephosphorylation(GO:0006470) |
| 0.0 |
0.9 |
GO:0008543 |
fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.0 |
0.1 |
GO:0018243 |
protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 |
0.2 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.0 |
0.0 |
GO:0006106 |
fumarate metabolic process(GO:0006106) |
| 0.0 |
0.4 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.0 |
0.3 |
GO:0006611 |
protein export from nucleus(GO:0006611) |
| 0.0 |
0.1 |
GO:0007254 |
JNK cascade(GO:0007254) |
| 0.0 |
0.1 |
GO:0099565 |
regulation of postsynaptic membrane potential(GO:0060078) excitatory postsynaptic potential(GO:0060079) chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.0 |
0.2 |
GO:0000910 |
cytokinesis(GO:0000910) |
| 0.0 |
0.0 |
GO:0017158 |
regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.0 |
0.2 |
GO:0007416 |
synapse assembly(GO:0007416) |
| 0.0 |
0.1 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 |
0.0 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.0 |
0.0 |
GO:0060748 |
tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 |
0.1 |
GO:0007506 |
gonadal mesoderm development(GO:0007506) |
| 0.0 |
0.1 |
GO:0045040 |
outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 |
0.1 |
GO:0051402 |
neuron apoptotic process(GO:0051402) |
| 0.0 |
0.1 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 |
0.1 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) histone H4-K12 acetylation(GO:0043983) |
| 0.0 |
0.0 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 |
0.0 |
GO:0033689 |
negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 |
0.1 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 |
0.1 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.0 |
0.6 |
GO:0032479 |
regulation of type I interferon production(GO:0032479) type I interferon production(GO:0032606) |
| 0.0 |
0.1 |
GO:0006893 |
Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 |
0.1 |
GO:0060644 |
mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 |
0.2 |
GO:0008333 |
endosome to lysosome transport(GO:0008333) |
| 0.0 |
0.0 |
GO:0060011 |
Sertoli cell proliferation(GO:0060011) |
| 0.0 |
0.1 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.0 |
GO:0010873 |
positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 |
0.2 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.0 |
0.0 |
GO:0046341 |
CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 |
0.0 |
GO:0007271 |
synaptic transmission, cholinergic(GO:0007271) |
| 0.0 |
0.0 |
GO:0006349 |
regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 |
0.1 |
GO:0006972 |
hyperosmotic response(GO:0006972) |
| 0.0 |
0.2 |
GO:0046129 |
purine nucleoside biosynthetic process(GO:0042451) purine ribonucleoside biosynthetic process(GO:0046129) |
| 0.0 |
0.1 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.0 |
GO:0006423 |
cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 |
0.1 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.2 |
GO:0045646 |
regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 |
0.0 |
GO:0070781 |
response to biotin(GO:0070781) |
| 0.0 |
0.2 |
GO:0031047 |
gene silencing by RNA(GO:0031047) |
| 0.0 |
0.6 |
GO:0050796 |
regulation of insulin secretion(GO:0050796) |
| 0.0 |
0.1 |
GO:0002076 |
osteoblast development(GO:0002076) |
| 0.0 |
0.1 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 |
0.1 |
GO:0060014 |
granulosa cell differentiation(GO:0060014) |
| 0.0 |
0.0 |
GO:0032392 |
DNA geometric change(GO:0032392) |
| 0.0 |
0.1 |
GO:0046485 |
ether lipid metabolic process(GO:0046485) |
| 0.0 |
0.1 |
GO:0010506 |
regulation of autophagy(GO:0010506) |
| 0.0 |
0.0 |
GO:0048840 |
otolith development(GO:0048840) |
| 0.0 |
0.2 |
GO:0038179 |
neurotrophin signaling pathway(GO:0038179) neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 |
0.0 |
GO:0034123 |
positive regulation of toll-like receptor signaling pathway(GO:0034123) |
| 0.0 |
0.1 |
GO:0007189 |
adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 |
0.2 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.1 |
GO:0071482 |
cellular response to light stimulus(GO:0071482) |
| 0.0 |
3.5 |
GO:0007283 |
spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 |
0.2 |
GO:0006101 |
citrate metabolic process(GO:0006101) tricarboxylic acid metabolic process(GO:0072350) |
| 0.0 |
0.0 |
GO:0070482 |
response to hypoxia(GO:0001666) response to decreased oxygen levels(GO:0036293) response to oxygen levels(GO:0070482) |
| 0.0 |
0.0 |
GO:0060968 |
regulation of gene silencing(GO:0060968) |
| 0.0 |
0.2 |
GO:0070664 |
negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) negative regulation of leukocyte proliferation(GO:0070664) |
| 0.0 |
0.1 |
GO:0035587 |
purinergic receptor signaling pathway(GO:0035587) purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 |
0.2 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.0 |
GO:1901798 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 |
0.0 |
GO:0002053 |
positive regulation of mesenchymal cell proliferation(GO:0002053) regulation of mesenchymal cell proliferation(GO:0010464) positive regulation of stem cell proliferation(GO:2000648) |
| 0.0 |
0.8 |
GO:0006814 |
sodium ion transport(GO:0006814) |
| 0.0 |
0.0 |
GO:0034339 |
obsolete regulation of transcription from RNA polymerase II promoter by nuclear hormone receptor(GO:0034339) |
| 0.0 |
2.4 |
GO:0008380 |
RNA splicing(GO:0008380) |
| 0.0 |
0.0 |
GO:0060317 |
cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 |
0.6 |
GO:0007608 |
sensory perception of smell(GO:0007608) |