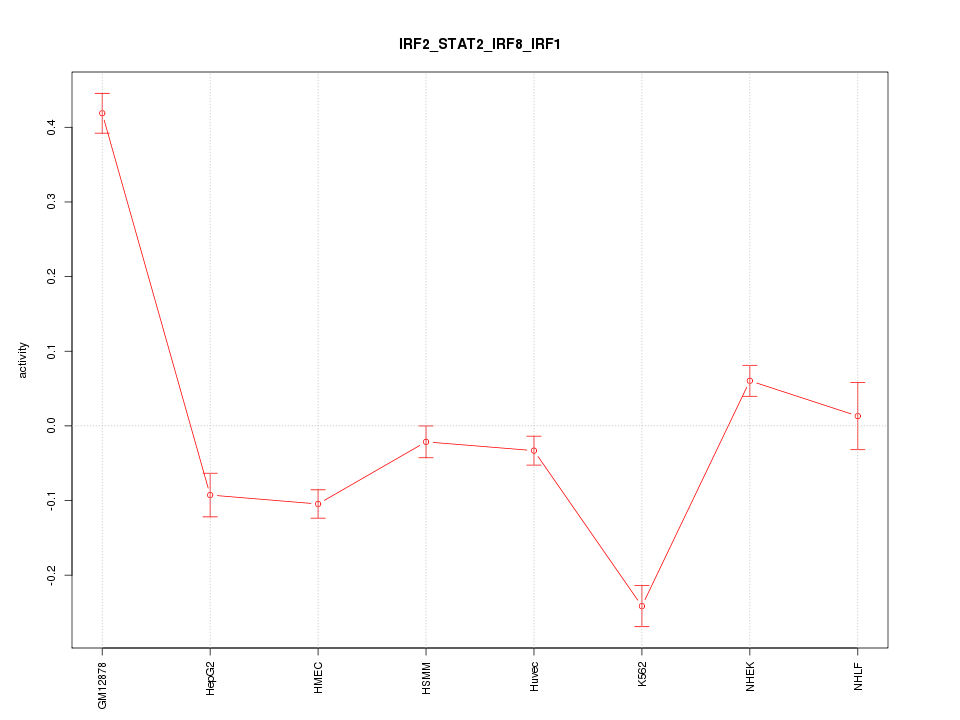
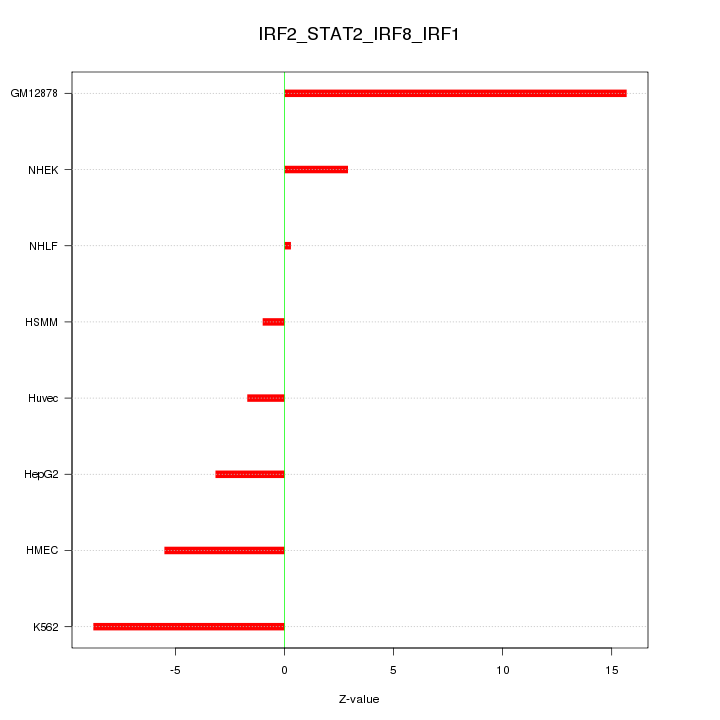
Motif ID: IRF2_STAT2_IRF8_IRF1
Z-value: 6.855
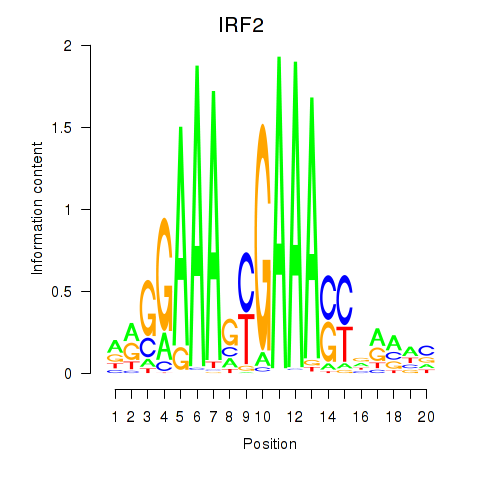
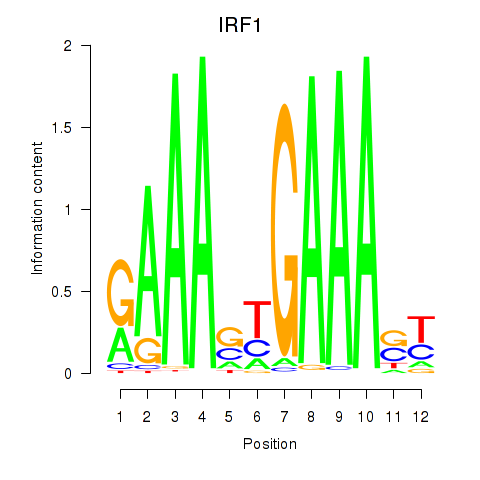
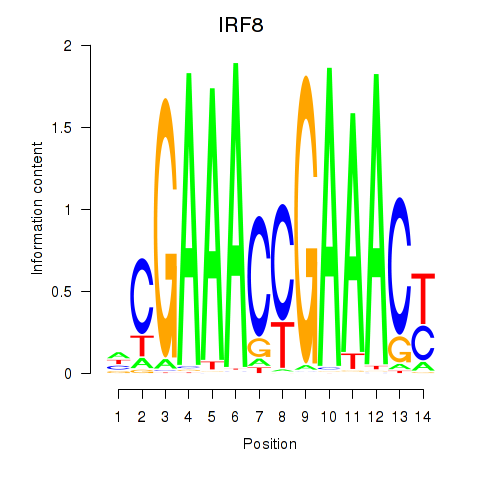
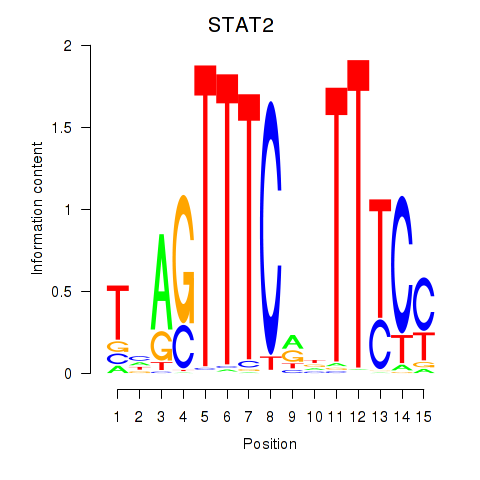
Transcription factors associated with IRF2_STAT2_IRF8_IRF1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| IRF1 | ENSG00000125347.9 | IRF1 |
| IRF2 | ENSG00000168310.6 | IRF2 |
| IRF8 | ENSG00000140968.6 | IRF8 |
| STAT2 | ENSG00000170581.9 | STAT2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 26.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 7.6 | 22.7 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 7.0 | 20.9 | GO:0045062 | extrathymic T cell differentiation(GO:0033078) extrathymic T cell selection(GO:0045062) |
| 5.6 | 28.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 2.6 | 7.8 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 2.6 | 7.7 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) purine nucleotide salvage(GO:0032261) nucleotide salvage(GO:0043173) inosine metabolic process(GO:0046102) xanthine metabolic process(GO:0046110) |
| 2.4 | 12.0 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 2.4 | 9.4 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 2.3 | 27.6 | GO:0032727 | positive regulation of interferon-alpha production(GO:0032727) |
| 2.0 | 184.1 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 2.0 | 9.9 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 1.7 | 18.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 1.6 | 29.2 | GO:0006906 | vesicle fusion(GO:0006906) |
| 1.6 | 19.4 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 1.5 | 6.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 1.5 | 5.9 | GO:0070842 | aggresome assembly(GO:0070842) |
| 1.4 | 7.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 1.4 | 43.7 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 1.4 | 5.4 | GO:1904037 | positive regulation of epithelial cell apoptotic process(GO:1904037) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 1.3 | 3.8 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 1.3 | 6.3 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 1.2 | 23.4 | GO:0070206 | protein trimerization(GO:0070206) |
| 1.2 | 34.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 1.1 | 25.3 | GO:0045730 | respiratory burst(GO:0045730) |
| 1.1 | 2.1 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 1.0 | 53.5 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 1.0 | 3.9 | GO:0051709 | regulation of killing of cells of other organism(GO:0051709) positive regulation of killing of cells of other organism(GO:0051712) |
| 1.0 | 6.8 | GO:0033034 | positive regulation of myeloid cell apoptotic process(GO:0033034) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.9 | 4.6 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.9 | 6.3 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.9 | 9.8 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.9 | 2.6 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.8 | 2.5 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.8 | 5.0 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.7 | 1.5 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.7 | 5.8 | GO:0050912 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.7 | 2.0 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.6 | 52.0 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.6 | 3.9 | GO:0015755 | fructose transport(GO:0015755) |
| 0.6 | 8.9 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.6 | 1.7 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.5 | 1.6 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.5 | 14.8 | GO:0006309 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.5 | 13.7 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.4 | 4.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.4 | 2.8 | GO:0045627 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.4 | 4.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.4 | 3.2 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.3 | 2.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.3 | 4.2 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.3 | 1.9 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.3 | 1.6 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.3 | 55.0 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.3 | 10.8 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.3 | 1.2 | GO:0039703 | single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.3 | 0.9 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.3 | 8.9 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.3 | 5.9 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.3 | 0.8 | GO:1903961 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.3 | 1.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.3 | 0.8 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.3 | 2.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 1.2 | GO:0090224 | regulation of spindle organization(GO:0090224) |
| 0.2 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.2 | 2.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 0.9 | GO:0071418 | uroporphyrinogen III biosynthetic process(GO:0006780) cellular response to amine stimulus(GO:0071418) |
| 0.2 | 0.7 | GO:0010273 | regulation of oxidative phosphorylation(GO:0002082) detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.2 | 0.9 | GO:0002902 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) |
| 0.2 | 1.3 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.2 | 2.8 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.2 | 0.2 | GO:0045060 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.2 | 1.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 1.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 4.0 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.2 | 4.0 | GO:0001906 | cell killing(GO:0001906) |
| 0.2 | 0.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 0.5 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.2 | 1.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.2 | 0.6 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.1 | 1.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.9 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 2.4 | GO:0002920 | regulation of humoral immune response(GO:0002920) |
| 0.1 | 6.0 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 0.6 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.1 | 0.8 | GO:0006184 | obsolete GTP catabolic process(GO:0006184) |
| 0.1 | 1.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 | 2.6 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.1 | 14.6 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.1 | 11.9 | GO:0042113 | B cell activation(GO:0042113) |
| 0.1 | 1.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 1.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.5 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 2.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.0 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.3 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.1 | 0.4 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 2.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.5 | GO:0032916 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.1 | 0.7 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 2.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 2.1 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 0.5 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.1 | GO:1903008 | organelle disassembly(GO:1903008) |
| 0.1 | 0.6 | GO:0051294 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.1 | 0.6 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.1 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.4 | GO:0016071 | mRNA metabolic process(GO:0016071) |
| 0.1 | 3.2 | GO:0097696 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.1 | 13.0 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.1 | 12.3 | GO:0009615 | response to virus(GO:0009615) |
| 0.1 | 8.8 | GO:0007281 | germ cell development(GO:0007281) |
| 0.1 | 0.3 | GO:2001259 | positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of cation channel activity(GO:2001259) |
| 0.1 | 0.1 | GO:0050942 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.1 | 0.2 | GO:0046546 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) male sex differentiation(GO:0046661) |
| 0.1 | 0.5 | GO:0032367 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.1 | 0.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 1.0 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.3 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.1 | 41.7 | GO:0006955 | immune response(GO:0006955) |
| 0.1 | 1.2 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 1.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.9 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.1 | 0.2 | GO:0072283 | mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003337) renal vesicle morphogenesis(GO:0072077) renal vesicle development(GO:0072087) cell differentiation involved in metanephros development(GO:0072202) metanephric nephron morphogenesis(GO:0072273) metanephric renal vesicle morphogenesis(GO:0072283) |
| 0.1 | 0.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.7 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 1.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) skeletal muscle organ development(GO:0060538) |
| 0.0 | 0.6 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 3.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 2.5 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 7.5 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 1.3 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.0 | 0.9 | GO:0050790 | regulation of catalytic activity(GO:0050790) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.4 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 2.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.5 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.4 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.7 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) regulation of mesenchymal cell proliferation(GO:0010464) |
| 0.0 | 1.8 | GO:0031668 | cellular response to extracellular stimulus(GO:0031668) |
| 0.0 | 1.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 1.0 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 2.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.4 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.4 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 3.3 | GO:0050900 | leukocyte migration(GO:0050900) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 2.5 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.8 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
| 0.0 | 0.5 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.2 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.0 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.2 | GO:0046854 | lipid phosphorylation(GO:0046834) phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.9 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 0.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0060711 | labyrinthine layer development(GO:0060711) |
| 0.0 | 1.9 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.3 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.8 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 3.7 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 6.5 | GO:0044419 | interspecies interaction between organisms(GO:0044419) |
| 0.0 | 1.0 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 1.0 | GO:0008154 | actin polymerization or depolymerization(GO:0008154) |
| 0.0 | 0.0 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.6 | GO:0061025 | membrane fusion(GO:0061025) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 40.5 | GO:0012505 | endomembrane system(GO:0012505) |
| 2.3 | 25.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 2.0 | 6.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 1.8 | 56.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 1.7 | 11.8 | GO:0042825 | TAP complex(GO:0042825) |
| 1.4 | 21.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 1.3 | 5.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 1.3 | 7.8 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.8 | 6.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.8 | 10.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.6 | 1.9 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) cytoplasmic side of membrane(GO:0098562) |
| 0.4 | 9.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.4 | 5.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.4 | 10.9 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.4 | 36.3 | GO:0016605 | PML body(GO:0016605) |
| 0.4 | 5.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.3 | 1.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 0.8 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 7.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 2.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 1.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 11.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 1.5 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 1.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 3.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 0.5 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 1.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 82.9 | GO:0005768 | endosome(GO:0005768) |
| 0.1 | 43.0 | GO:0005792 | obsolete microsome(GO:0005792) |
| 0.1 | 6.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 5.6 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.2 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.1 | 17.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.3 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 21.7 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.1 | 2.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 1.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 17.2 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 0.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 3.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 3.0 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 0.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 2.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 0.6 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.1 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 6.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 0.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 7.1 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.1 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 1.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 43.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.6 | GO:0035097 | methyltransferase complex(GO:0034708) histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 13.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) nuclear outer membrane-endoplasmic reticulum membrane network(GO:0042175) |
| 0.0 | 0.6 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 74.0 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 1.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.7 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 2.2 | GO:0005795 | Golgi stack(GO:0005795) organelle subcompartment(GO:0031984) Golgi subcompartment(GO:0098791) |
| 0.0 | 0.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 155.1 | GO:0005622 | intracellular(GO:0005622) |
| 0.0 | 0.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:0030425 | dendrite(GO:0030425) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 26.4 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) exoribonuclease II activity(GO:0008859) |
| 4.6 | 13.9 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 2.9 | 11.6 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 2.6 | 46.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 2.4 | 7.2 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 1.9 | 24.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 1.9 | 9.4 | GO:0046980 | oligopeptide-transporting ATPase activity(GO:0015421) tapasin binding(GO:0046980) |
| 1.9 | 56.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 1.8 | 5.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.7 | 5.0 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 1.4 | 11.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 1.4 | 6.9 | GO:0005534 | galactose binding(GO:0005534) |
| 1.3 | 8.9 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 1.2 | 6.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 1.2 | 2.4 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 1.1 | 16.5 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 1.1 | 6.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 1.0 | 3.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 1.0 | 14.8 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 1.0 | 6.8 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.9 | 4.6 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) |
| 0.9 | 8.7 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.8 | 2.5 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.7 | 65.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.7 | 5.9 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.7 | 3.9 | GO:0019863 | IgE binding(GO:0019863) |
| 0.6 | 7.8 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.6 | 7.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.6 | 3.9 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.6 | 4.4 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.5 | 3.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.5 | 18.2 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.5 | 13.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.5 | 1.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.4 | 5.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.4 | 5.7 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.4 | 1.7 | GO:0008907 | integrase activity(GO:0008907) |
| 0.4 | 3.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.4 | 1.2 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.4 | 4.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.3 | 1.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.3 | 35.5 | GO:0003704 | obsolete specific RNA polymerase II transcription factor activity(GO:0003704) |
| 0.3 | 0.9 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.3 | 1.8 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.3 | 1.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 4.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 43.8 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.3 | 30.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.3 | 11.8 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.2 | 1.2 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.2 | 32.9 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.2 | 0.9 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.2 | 1.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.2 | 1.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.2 | 62.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.2 | 7.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 1.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.2 | 1.8 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 0.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 2.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 1.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 2.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.2 | 0.7 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 1.3 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.2 | 0.5 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.2 | 0.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 2.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 2.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 1.0 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 0.6 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 21.6 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 0.5 | GO:0034701 | tripeptidase activity(GO:0034701) |
| 0.1 | 1.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 1.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 1.7 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.1 | 5.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 5.2 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.1 | 2.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.6 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 1.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.1 | 0.4 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.1 | 0.9 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.1 | 3.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 2.8 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.2 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.3 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 10.0 | GO:0003702 | obsolete RNA polymerase II transcription factor activity(GO:0003702) |
| 0.1 | 0.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.1 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.1 | 1.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 1.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 4.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 2.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.6 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 12.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.8 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 3.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 31.6 | GO:0003700 | transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 41.2 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.3 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 2.2 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.6 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 1.0 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 0.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 19.4 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
| 0.8 | 20.5 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.4 | 2.9 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.4 | 8.5 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.4 | 21.5 | SIG_PIP3_SIGNALING_IN_B_LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 3.1 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 4.5 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.2 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 7.0 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 1.3 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 0.9 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |