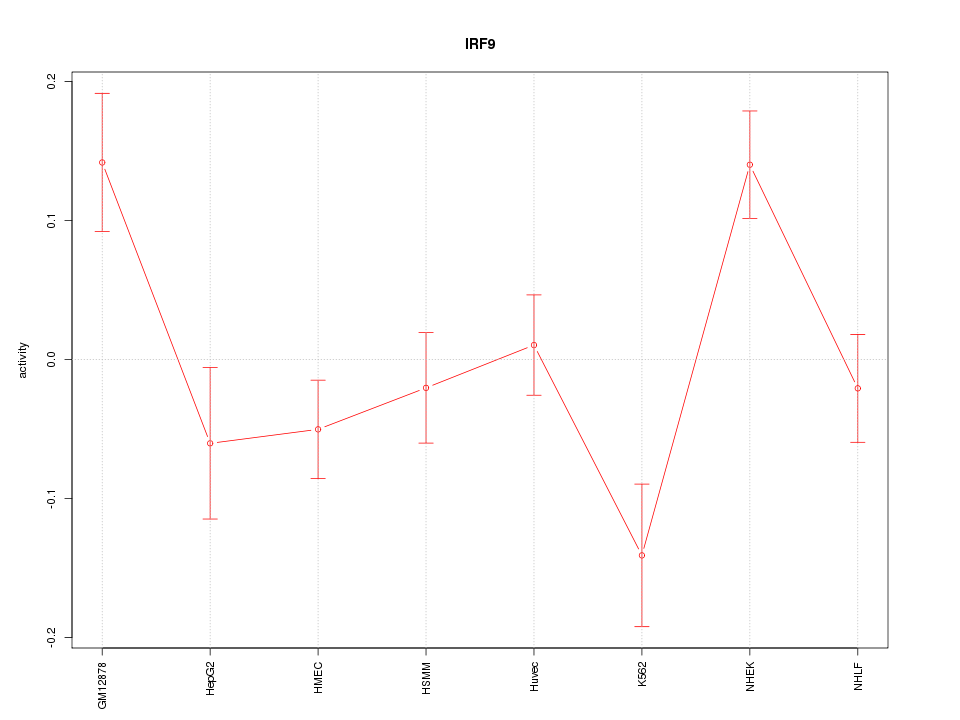
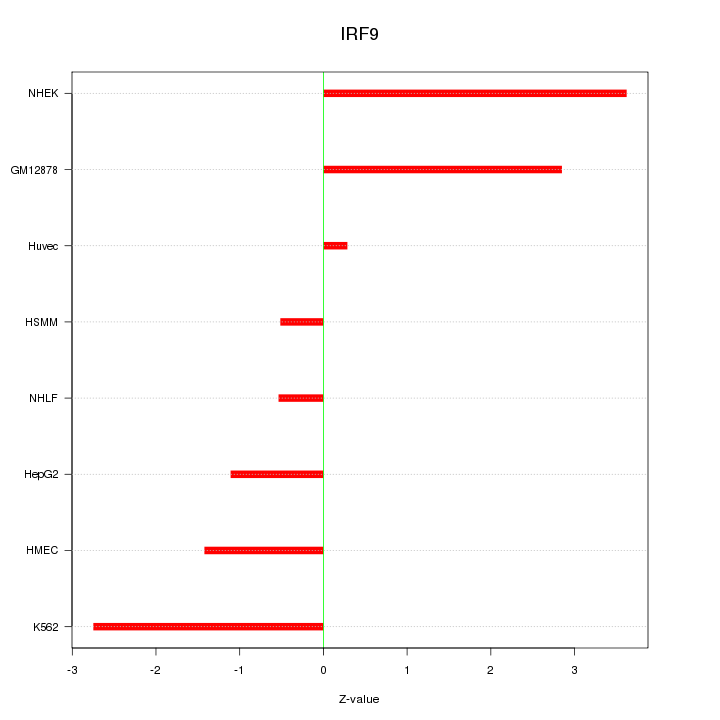
Motif ID: IRF9
Z-value: 2.021
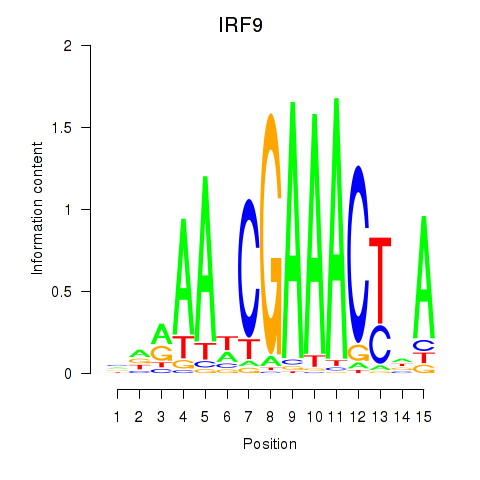
Transcription factors associated with IRF9:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| IRF9 | ENSG00000213928.4 | IRF9 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.7 | 4.8 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.6 | 7.3 | GO:0032727 | positive regulation of interferon-alpha production(GO:0032727) |
| 0.6 | 1.7 | GO:0002431 | Fc receptor mediated stimulatory signaling pathway(GO:0002431) regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.4 | 2.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.4 | 1.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.4 | 36.8 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.3 | 8.8 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.3 | 1.7 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.2 | 0.6 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.2 | 1.9 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 7.6 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.1 | 2.3 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 0.3 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 1.9 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 1.4 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 0.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 0.4 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.9 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.7 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.4 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.5 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 2.0 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.0 | 0.7 | GO:0097696 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 | 2.6 | GO:0009615 | response to virus(GO:0009615) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 12.8 | GO:0005792 | obsolete microsome(GO:0005792) |
| 0.0 | 1.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 4.7 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 4.5 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 1.7 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 12.8 | GO:0005654 | nucleoplasm(GO:0005654) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.7 | 4.8 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.2 | 2.9 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.2 | 1.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.7 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 1.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.4 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) |
| 0.1 | 10.0 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.1 | 2.9 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.1 | 0.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.1 | 5.4 | GO:0003704 | obsolete specific RNA polymerase II transcription factor activity(GO:0003704) |
| 0.1 | 8.8 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.0 | 12.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 4.9 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 5.1 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 3.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 1.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.5 | GO:0004386 | helicase activity(GO:0004386) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.8 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.9 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 3.1 | SIG_PIP3_SIGNALING_IN_B_LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 2.3 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |