Motif ID: ISL1
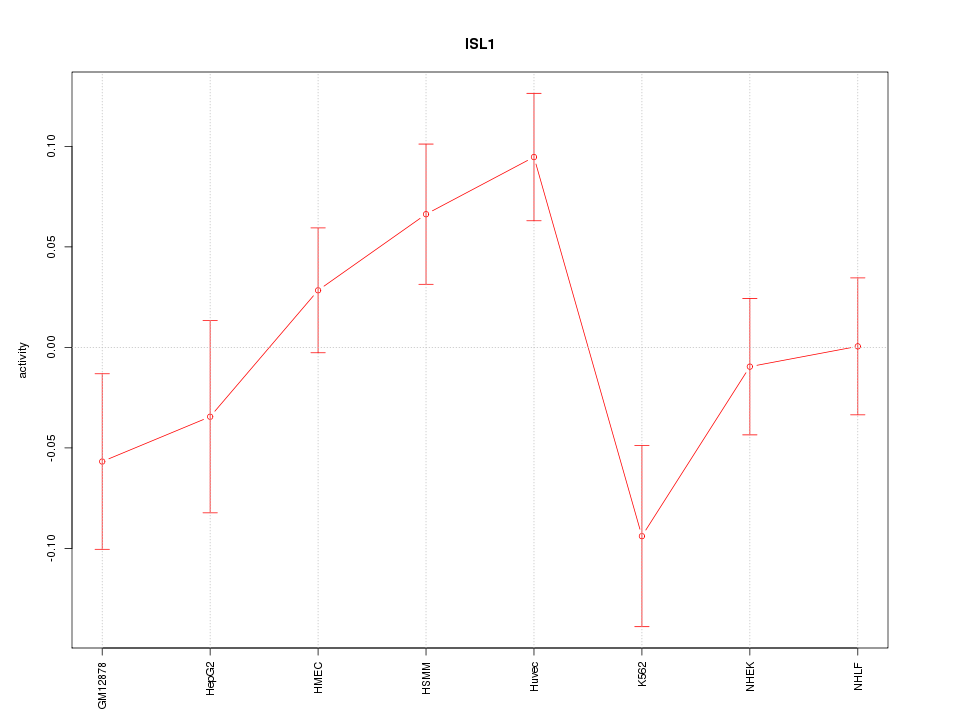
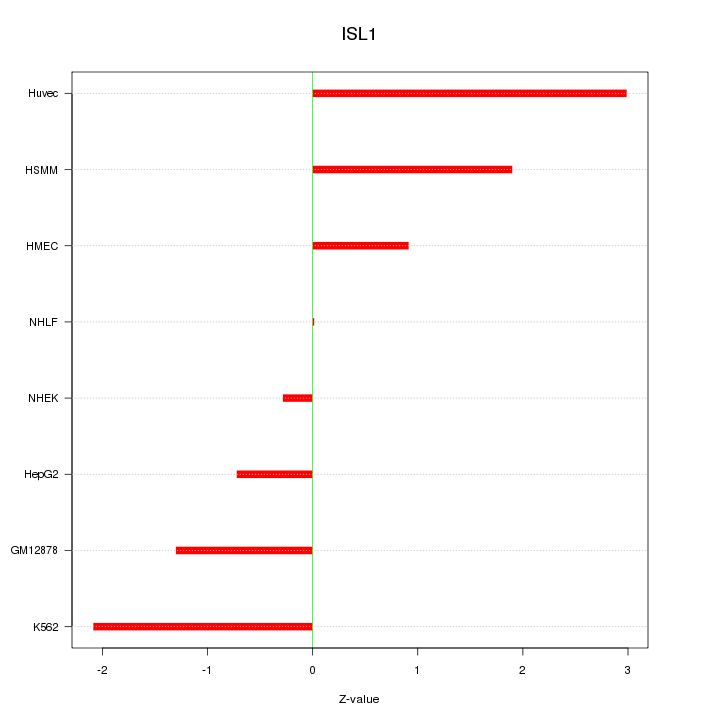
Z-value: 1.582
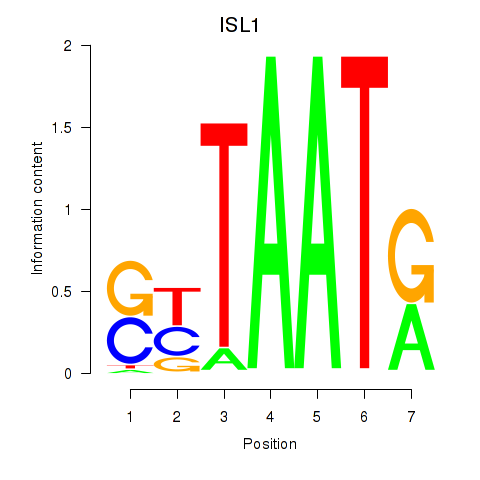
Transcription factors associated with ISL1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| ISL1 | ENSG00000016082.10 | ISL1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 7.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.5 | 3.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.2 | 1.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.2 | 1.2 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.2 | 1.6 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.2 | 1.6 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.2 | 1.3 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.2 | 0.6 | GO:0050653 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.2 | 0.6 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 0.3 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 2.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.2 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.1 | 0.4 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.2 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 | 0.3 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.7 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.1 | 0.2 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.8 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.2 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 5.1 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.6 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 | 3.3 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 1.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.3 | GO:0001823 | ureteric bud development(GO:0001657) mesonephros development(GO:0001823) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.0 | 0.2 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 3.9 | GO:0030336 | negative regulation of cell migration(GO:0030336) negative regulation of cell motility(GO:2000146) |
| 0.0 | 0.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.0 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 1.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.0 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 5.0 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.6 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 2.4 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 1.0 | GO:0034249 | negative regulation of translation(GO:0017148) negative regulation of cellular amide metabolic process(GO:0034249) |
| 0.0 | 4.5 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.2 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 1.3 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.8 | GO:0007202 | activation of phospholipase C activity(GO:0007202) positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.0 | 6.2 | GO:0007010 | cytoskeleton organization(GO:0007010) |
| 0.0 | 1.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.3 | 0.8 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 1.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 0.7 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 5.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 4.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 7.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 1.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.5 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.3 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 2.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0030286 | dynein complex(GO:0030286) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 3.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.3 | 3.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.3 | 3.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 0.7 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.2 | 0.6 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.2 | 1.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 1.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 4.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.6 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 4.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.1 | 1.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 4.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.6 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.2 | GO:0017163 | obsolete basal transcription repressor activity(GO:0017163) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 7.8 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 2.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 3.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |