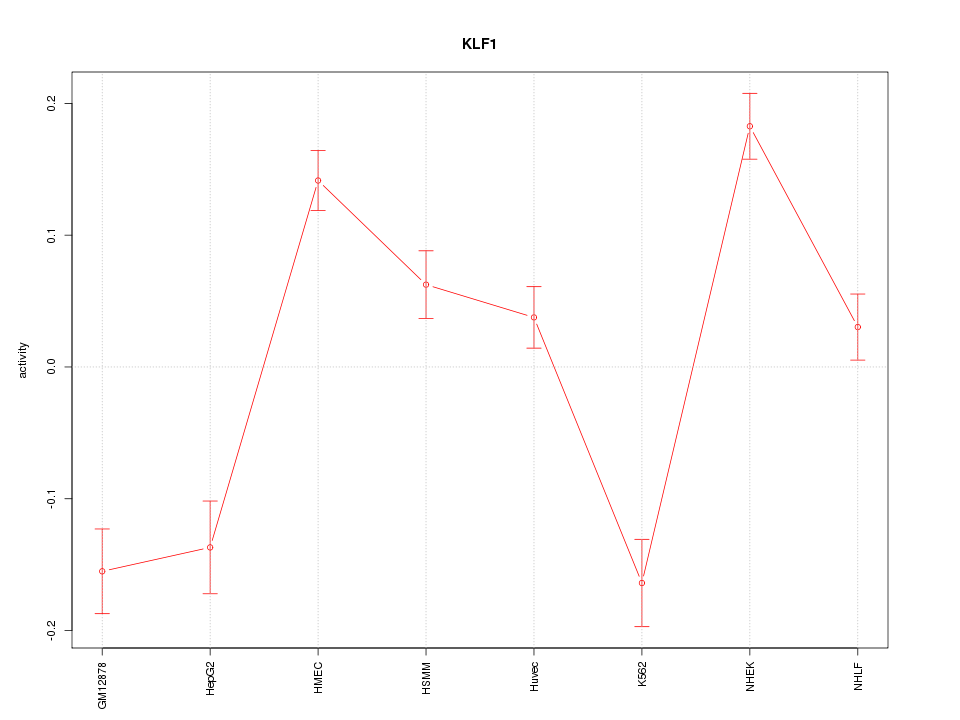
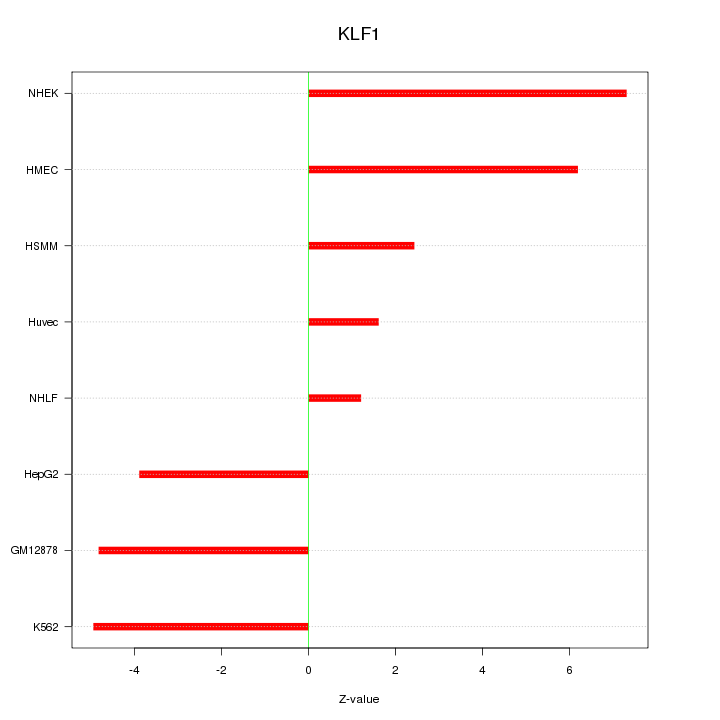
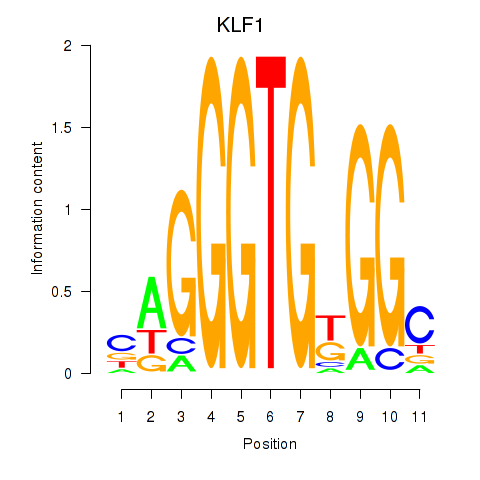
Motif ID: KLF1
Z-value: 4.537
Transcription factors associated with KLF1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| KLF1 | ENSG00000105610.4 | KLF1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 1.7 | 5.1 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 1.5 | 4.5 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 1.5 | 5.8 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 1.4 | 4.3 | GO:0048627 | myoblast development(GO:0048627) |
| 1.3 | 37.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 1.3 | 11.5 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 1.3 | 8.8 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 1.0 | 2.9 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.8 | 2.5 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.8 | 2.3 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.8 | 3.8 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.7 | 2.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.7 | 2.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.7 | 4.0 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.6 | 16.8 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.6 | 1.8 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.6 | 3.9 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.6 | 3.4 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.6 | 4.5 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.5 | 1.6 | GO:0071603 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) endothelial cell-cell adhesion(GO:0071603) |
| 0.5 | 2.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.5 | 2.9 | GO:2000124 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.5 | 1.5 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.5 | 8.2 | GO:0071333 | cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.4 | 6.6 | GO:0051927 | obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.4 | 1.3 | GO:0001300 | chronological cell aging(GO:0001300) detection of hypoxia(GO:0070483) |
| 0.4 | 2.5 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.4 | 1.2 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) post-embryonic camera-type eye development(GO:0031077) |
| 0.4 | 4.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.4 | 2.7 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.4 | 1.5 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.4 | 1.5 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.3 | 1.4 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.3 | 1.7 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.3 | 6.7 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.3 | 1.7 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.3 | 5.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.3 | 1.1 | GO:1902116 | negative regulation of organelle assembly(GO:1902116) |
| 0.3 | 2.0 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.3 | 2.5 | GO:0019359 | nicotinamide nucleotide biosynthetic process(GO:0019359) |
| 0.2 | 0.7 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.2 | 5.0 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.2 | 3.8 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative chemotaxis(GO:0050919) |
| 0.2 | 2.5 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.2 | 0.2 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.2 | 2.7 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.2 | 1.8 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.2 | 4.0 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.2 | 6.5 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.2 | 0.8 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.2 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 2.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 1.2 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 | 1.3 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.1 | 1.6 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 0.8 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.4 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 2.4 | GO:0007628 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.1 | 0.6 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 9.3 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 4.9 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.1 | 0.8 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 2.5 | GO:0060749 | mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.1 | 3.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.6 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 1.0 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.3 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 1.3 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 3.6 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 5.6 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.1 | 0.6 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 2.0 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.1 | 2.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.8 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 0.6 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.7 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 0.4 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.1 | 1.0 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.1 | 1.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 1.8 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 0.9 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 1.1 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 0.2 | GO:0034337 | RNA folding(GO:0034337) DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 1.7 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 5.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 2.6 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 4.8 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 3.4 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.5 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 1.9 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.2 | GO:0090343 | positive regulation of cell aging(GO:0090343) |
| 0.0 | 0.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 5.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.7 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 4.7 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 5.2 | GO:0071466 | xenobiotic metabolic process(GO:0006805) cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 2.4 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 1.4 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 1.0 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.4 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.8 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 2.4 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.7 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 1.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 1.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 2.4 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 2.9 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.7 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.2 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.6 | GO:0051149 | positive regulation of muscle cell differentiation(GO:0051149) |
| 0.0 | 0.9 | GO:0046822 | regulation of nucleocytoplasmic transport(GO:0046822) |
| 0.0 | 0.8 | GO:0007292 | female gamete generation(GO:0007292) |
| 0.0 | 0.0 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 17.0 | GO:0001527 | microfibril(GO:0001527) |
| 1.0 | 5.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.8 | 11.6 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.7 | 2.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.6 | 4.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.6 | 16.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.5 | 3.8 | GO:0043205 | fibril(GO:0043205) |
| 0.5 | 23.5 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.4 | 1.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) PAS complex(GO:0070772) |
| 0.4 | 1.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.3 | 1.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.3 | 1.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.3 | 1.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 1.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 20.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 4.3 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.2 | 0.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 0.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 1.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 2.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 1.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 0.6 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.2 | 2.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 2.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 12.7 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 4.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.1 | 0.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 9.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 6.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 2.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 2.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 19.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 0.6 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 2.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.1 | 1.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.7 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 4.5 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.1 | 0.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 6.9 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 1.0 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 1.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.4 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.0 | 1.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 2.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 3.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 5.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.6 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 9.0 | GO:0005792 | obsolete microsome(GO:0005792) |
| 0.0 | 1.2 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 12.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.6 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 1.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 29.1 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 1.3 | 2.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 1.1 | 3.4 | GO:0048184 | obsolete follistatin binding(GO:0048184) |
| 1.1 | 6.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.1 | 8.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.9 | 4.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.9 | 5.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.9 | 29.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.8 | 3.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.8 | 3.8 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.7 | 7.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.6 | 5.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.6 | 2.5 | GO:0004473 | malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.6 | 1.8 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.5 | 15.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.5 | 2.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.5 | 1.5 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.4 | 1.3 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.4 | 2.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.4 | 0.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.4 | 1.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 4.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.3 | 12.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.3 | 4.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 0.8 | GO:0050062 | long-chain-fatty-acyl-CoA reductase activity(GO:0050062) |
| 0.3 | 1.3 | GO:0005534 | galactose binding(GO:0005534) |
| 0.3 | 1.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.3 | 0.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.3 | 5.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 4.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 2.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 2.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 0.9 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.2 | 2.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 1.8 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.2 | 4.1 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.2 | 0.7 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.2 | 2.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 2.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 20.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 1.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 1.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 5.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 15.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 1.0 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 2.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.1 | 0.6 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.1 | 2.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 4.0 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.1 | 1.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 2.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.6 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 19.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 3.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 1.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 6.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.0 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.1 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.5 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.3 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
| 0.1 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.6 | GO:0004835 | tubulin-tyrosine ligase activity(GO:0004835) |
| 0.1 | 0.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 10.2 | GO:0003702 | obsolete RNA polymerase II transcription factor activity(GO:0003702) |
| 0.1 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.7 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 1.8 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 20.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 5.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 6.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 1.6 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.8 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.9 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 8.2 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0016566 | obsolete specific transcriptional repressor activity(GO:0016566) |
| 0.0 | 0.4 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 10.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 2.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 6.2 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 4.1 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.1 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.3 | 17.1 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
| 0.2 | 17.5 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.1 | 2.8 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 3.8 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.1 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 1.2 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.7 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | ST_STAT3_PATHWAY | STAT3 Pathway |
| 0.0 | 0.8 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |