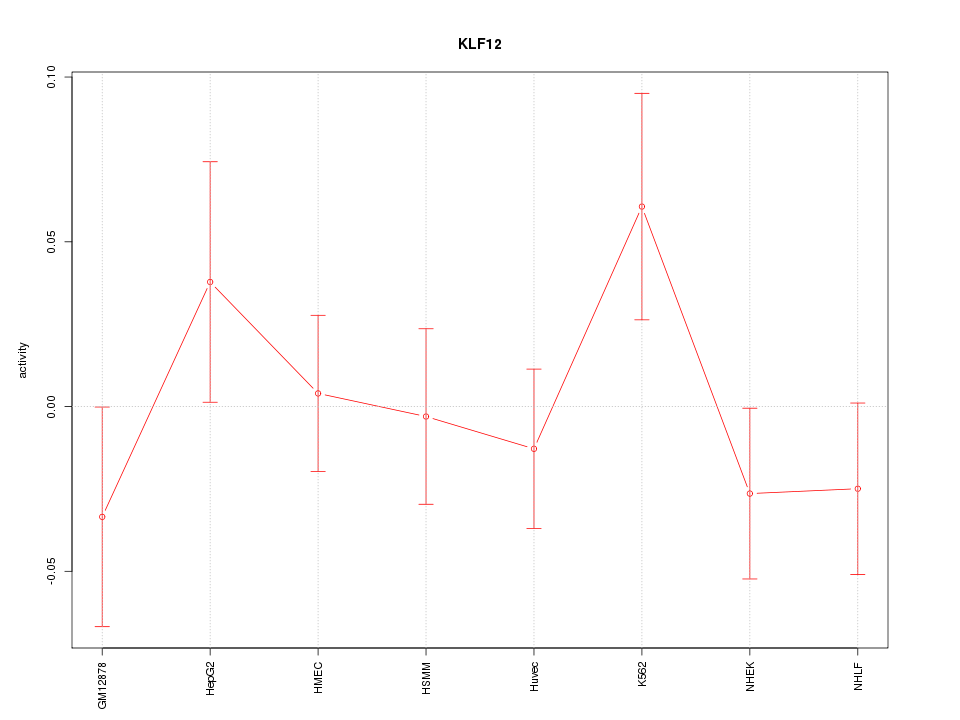
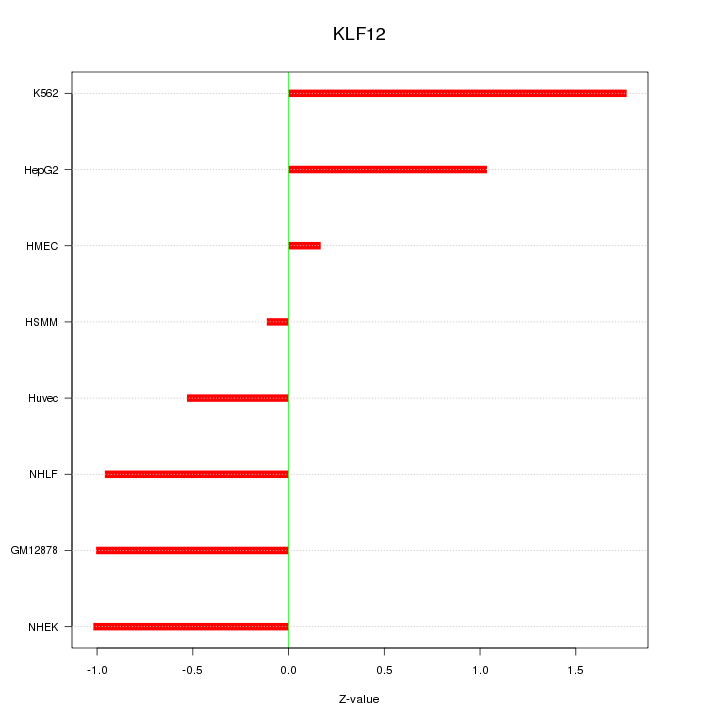
Motif ID: KLF12
Z-value: 0.967
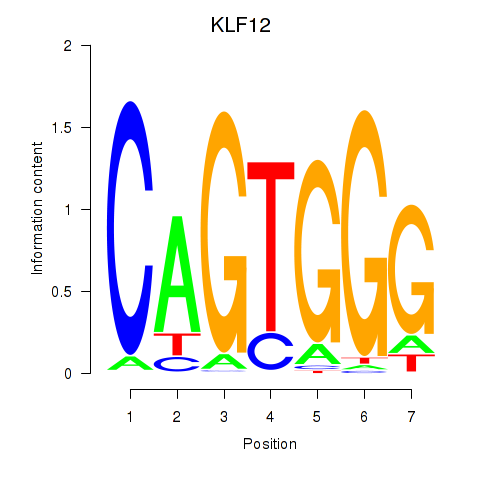
Transcription factors associated with KLF12:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| KLF12 | ENSG00000118922.12 | KLF12 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.6 | 3.1 | GO:0007614 | short-term memory(GO:0007614) |
| 0.5 | 1.0 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.4 | 1.3 | GO:0055005 | optic placode formation involved in camera-type eye formation(GO:0046619) ventricular cardiac myofibril assembly(GO:0055005) regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.4 | 1.2 | GO:0031077 | post-embryonic camera-type eye development(GO:0031077) Spemann organizer formation(GO:0060061) |
| 0.3 | 0.9 | GO:0060896 | neural plate anterior/posterior regionalization(GO:0021999) neural plate pattern specification(GO:0060896) neural plate regionalization(GO:0060897) |
| 0.3 | 1.1 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.2 | 0.7 | GO:1901724 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.2 | 0.7 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) protein hexamerization(GO:0034214) |
| 0.2 | 1.0 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 0.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 0.6 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.8 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 0.9 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.1 | 0.4 | GO:0061198 | fungiform papilla development(GO:0061196) fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 0.1 | 0.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.2 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.1 | 0.3 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 1.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.7 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.1 | 0.9 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.4 | GO:0043247 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.1 | 0.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.2 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.1 | 1.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.4 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 0.2 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.1 | 0.3 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 1.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.3 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.1 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.1 | GO:0046520 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.3 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.2 | GO:0045953 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) negative regulation of natural killer cell mediated immunity(GO:0002716) negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.0 | 0.6 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.3 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.4 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.2 | GO:0090343 | positive regulation of cell aging(GO:0090343) |
| 0.0 | 0.5 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.4 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.2 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.4 | GO:0034372 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 1.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.0 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.0 | 0.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.1 | 1.0 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.4 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.5 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 2.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 1.7 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 0.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.6 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 1.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.4 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.3 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.3 | GO:0016731 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 1.4 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.9 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.4 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 1.9 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.1 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0015187 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) glycine transmembrane transporter activity(GO:0015187) glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 1.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 1.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 3.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.9 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.6 | GO:0034979 | NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.3 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 1.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.3 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST_ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.8 | ST_DIFFERENTIATION_PATHWAY_IN_PC12_CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |