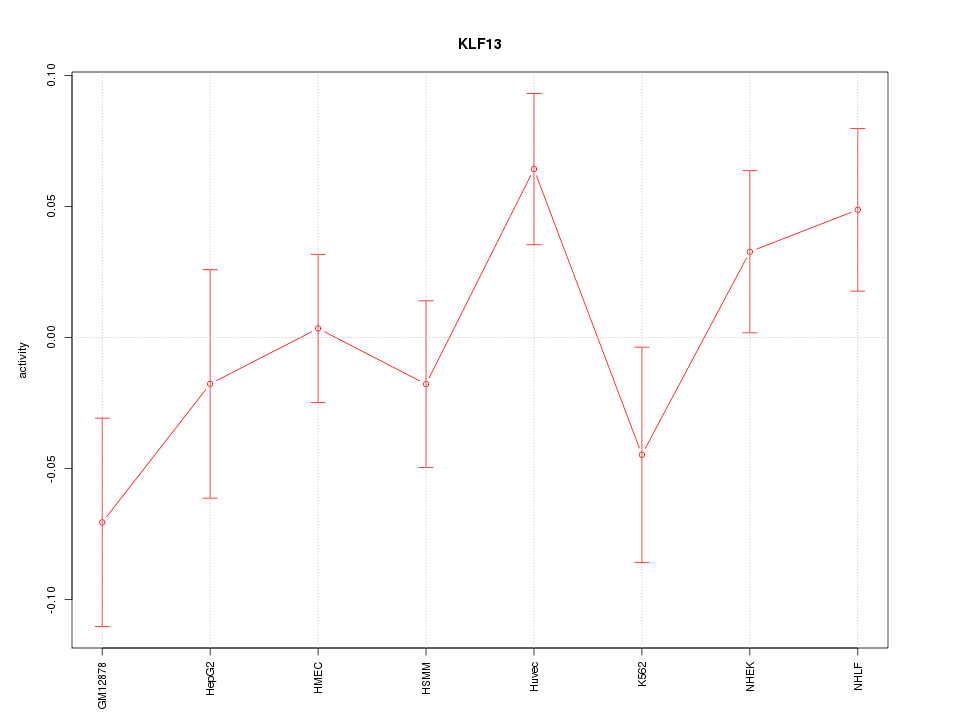
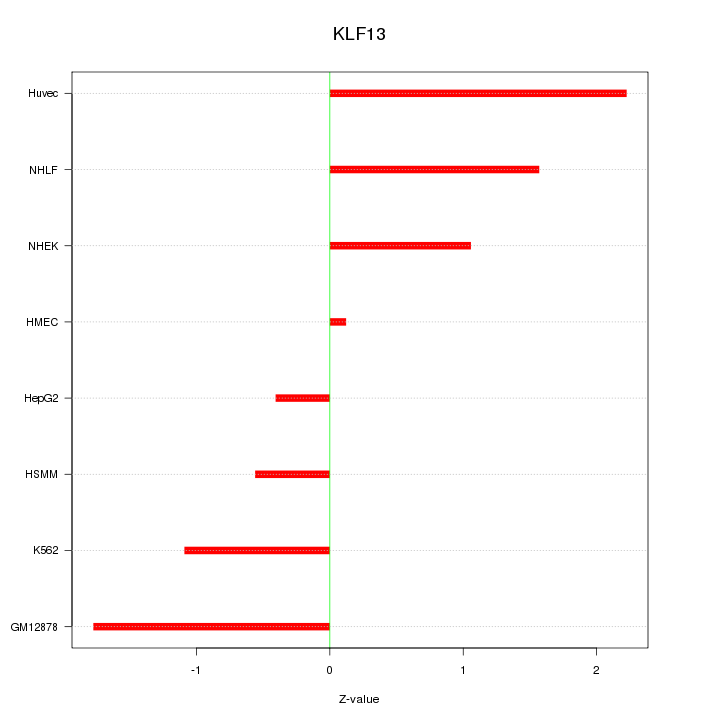
Motif ID: KLF13
Z-value: 1.292
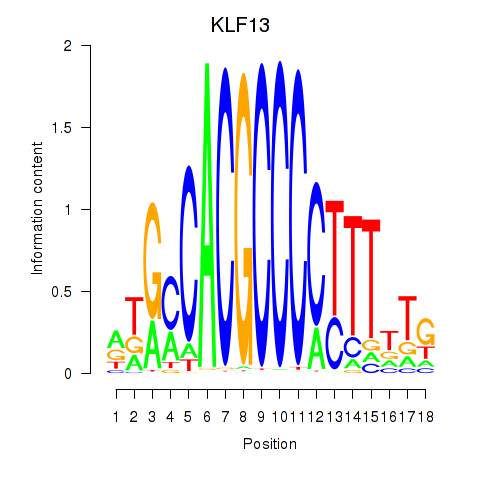
Transcription factors associated with KLF13:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| KLF13 | ENSG00000169926.5 | KLF13 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.5 | 1.5 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.3 | 0.9 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.3 | 3.8 | GO:0000052 | citrulline metabolic process(GO:0000052) arginine catabolic process(GO:0006527) |
| 0.3 | 0.8 | GO:0003011 | diaphragm contraction(GO:0002086) involuntary skeletal muscle contraction(GO:0003011) regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.2 | 0.7 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.2 | 0.8 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 2.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 0.4 | GO:0006740 | glyoxylate cycle(GO:0006097) NADPH regeneration(GO:0006740) |
| 0.1 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.3 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 1.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 0.8 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.6 | GO:0051294 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.1 | 0.6 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.5 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.1 | 0.5 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.3 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 1.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.5 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.7 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.4 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 3.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.1 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.3 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 1.8 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.8 | GO:0042384 | cilium assembly(GO:0042384) |
| 0.0 | 0.1 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.3 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 1.4 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 0.2 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.6 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 0.8 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 0.8 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 2.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 3.1 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.2 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 1.4 | GO:0005625 | obsolete soluble fraction(GO:0005625) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 1.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.5 | 3.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.5 | 1.5 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.3 | 0.8 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.2 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 0.5 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.2 | 0.8 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.4 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 1.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.5 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.8 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.3 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.1 | 0.4 | GO:0052658 | PH domain binding(GO:0042731) inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.1 | 2.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 1.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 3.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.6 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.5 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.6 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.9 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0032451 | demethylase activity(GO:0032451) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.1 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |