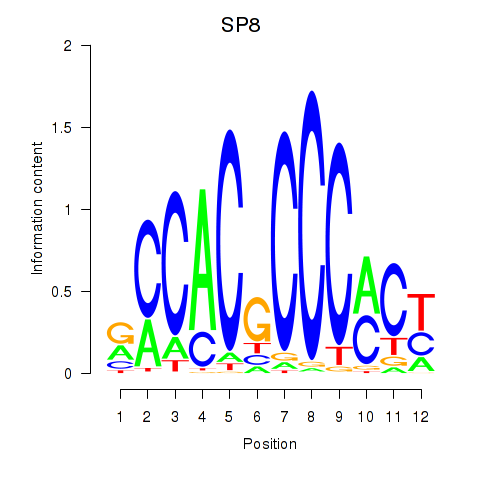
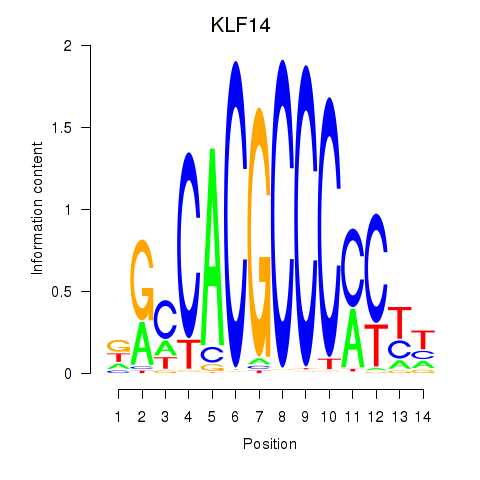
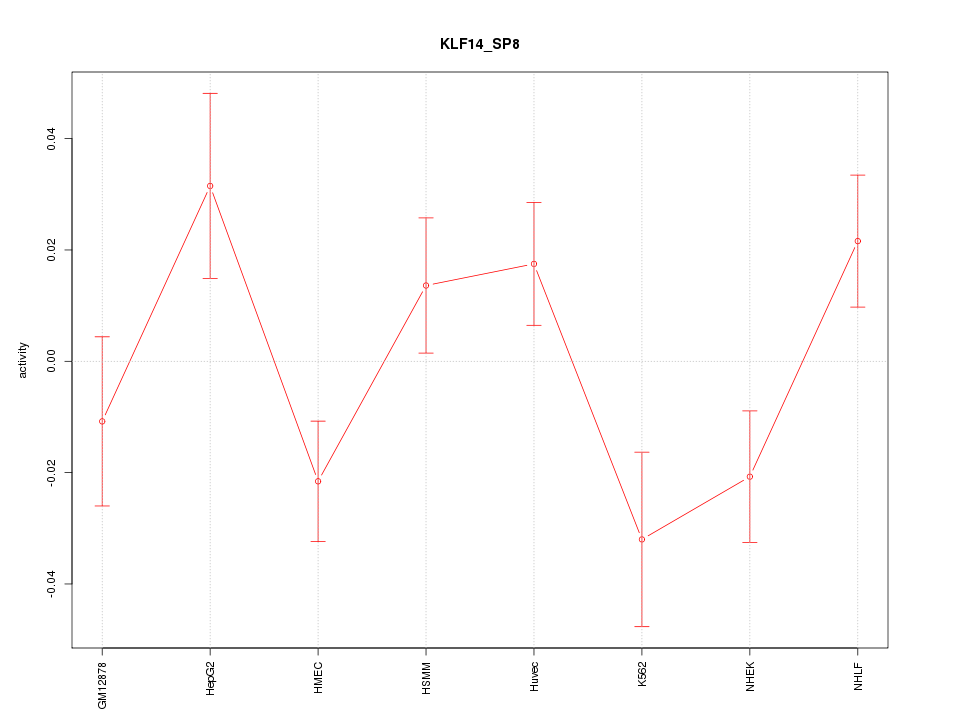
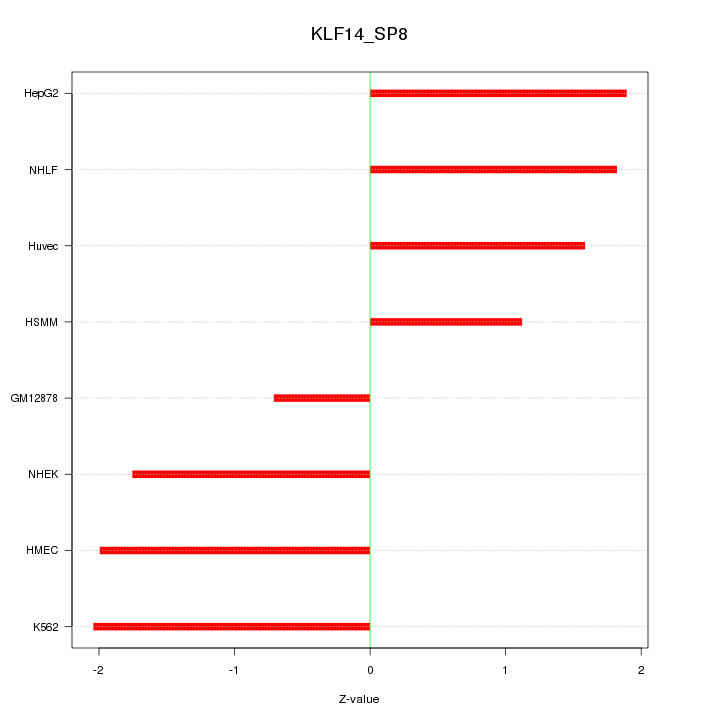
| 1.0 |
2.9 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 1.0 |
3.8 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.8 |
3.4 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.7 |
2.0 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.6 |
1.9 |
GO:0046210 |
nitric oxide catabolic process(GO:0046210) |
| 0.6 |
1.8 |
GO:0010757 |
chronological cell aging(GO:0001300) arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) negative regulation of plasminogen activation(GO:0010757) negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.6 |
1.7 |
GO:0043371 |
negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.5 |
1.5 |
GO:0018377 |
N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.4 |
1.3 |
GO:0002082 |
regulation of oxidative phosphorylation(GO:0002082) |
| 0.4 |
5.9 |
GO:0000052 |
citrulline metabolic process(GO:0000052) |
| 0.4 |
1.1 |
GO:0006051 |
mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.4 |
1.4 |
GO:0008065 |
establishment of blood-nerve barrier(GO:0008065) |
| 0.3 |
3.4 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.3 |
2.0 |
GO:0006011 |
UDP-glucose metabolic process(GO:0006011) UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.3 |
0.7 |
GO:0060059 |
embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.3 |
2.7 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.3 |
1.5 |
GO:0019255 |
glucose 1-phosphate metabolic process(GO:0019255) |
| 0.3 |
0.9 |
GO:0061298 |
progesterone secretion(GO:0042701) retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.3 |
0.6 |
GO:0010273 |
detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.3 |
1.1 |
GO:0048619 |
embryonic hindgut morphogenesis(GO:0048619) |
| 0.3 |
0.8 |
GO:0009756 |
carbohydrate mediated signaling(GO:0009756) |
| 0.3 |
1.8 |
GO:0060414 |
aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 |
0.6 |
GO:0042339 |
keratan sulfate metabolic process(GO:0042339) |
| 0.2 |
0.8 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 |
1.0 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 |
0.6 |
GO:0035234 |
ectopic germ cell programmed cell death(GO:0035234) |
| 0.2 |
1.6 |
GO:0006072 |
glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.2 |
0.5 |
GO:0045198 |
establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.2 |
1.1 |
GO:0030186 |
melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.2 |
0.7 |
GO:0010900 |
regulation of phosphatidylcholine catabolic process(GO:0010899) negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.2 |
1.2 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.2 |
1.7 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.2 |
0.5 |
GO:0030205 |
dermatan sulfate metabolic process(GO:0030205) dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 |
0.6 |
GO:0043441 |
acetoacetic acid biosynthetic process(GO:0043441) |
| 0.1 |
0.4 |
GO:1904358 |
positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.1 |
0.4 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 |
1.0 |
GO:0003197 |
endocardial cushion development(GO:0003197) |
| 0.1 |
0.8 |
GO:0019673 |
GDP-mannose metabolic process(GO:0019673) |
| 0.1 |
0.3 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.1 |
2.7 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.1 |
0.3 |
GO:0090206 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 |
0.7 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.1 |
0.1 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 |
0.4 |
GO:0046520 |
diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.1 |
0.6 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.1 |
0.7 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.1 |
0.2 |
GO:0071786 |
endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 |
0.5 |
GO:0034442 |
plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.1 |
0.6 |
GO:0046642 |
negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 |
0.3 |
GO:0032119 |
sequestering of zinc ion(GO:0032119) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 |
0.1 |
GO:0070997 |
neuron apoptotic process(GO:0051402) neuron death(GO:0070997) |
| 0.1 |
1.1 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 |
0.1 |
GO:0006903 |
vesicle targeting(GO:0006903) |
| 0.1 |
0.5 |
GO:0006651 |
diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 |
0.3 |
GO:0035608 |
protein deglutamylation(GO:0035608) |
| 0.1 |
0.3 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.1 |
0.8 |
GO:0070296 |
sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.1 |
0.3 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.1 |
0.4 |
GO:0045048 |
protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 |
0.6 |
GO:0050665 |
hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.1 |
0.3 |
GO:1901419 |
regulation of response to alcohol(GO:1901419) |
| 0.1 |
0.1 |
GO:0010882 |
regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.1 |
0.4 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.1 |
0.3 |
GO:0050758 |
thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 |
0.6 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.3 |
GO:0031077 |
post-embryonic camera-type eye development(GO:0031077) |
| 0.1 |
0.6 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
0.2 |
GO:0052805 |
histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 |
0.6 |
GO:0022417 |
protein maturation by protein folding(GO:0022417) |
| 0.1 |
0.3 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 |
0.2 |
GO:0034244 |
negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 |
0.6 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.1 |
0.8 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 |
0.6 |
GO:0032075 |
positive regulation of nuclease activity(GO:0032075) |
| 0.1 |
0.4 |
GO:0000050 |
urea cycle(GO:0000050) |
| 0.1 |
0.2 |
GO:0034036 |
purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 |
0.2 |
GO:0060596 |
mammary placode formation(GO:0060596) |
| 0.1 |
0.2 |
GO:0002575 |
basophil chemotaxis(GO:0002575) |
| 0.1 |
0.3 |
GO:0015825 |
L-serine transport(GO:0015825) |
| 0.1 |
0.2 |
GO:0006740 |
NADPH regeneration(GO:0006740) |
| 0.1 |
0.4 |
GO:1903008 |
organelle disassembly(GO:1903008) |
| 0.1 |
0.2 |
GO:0033578 |
protein glycosylation in Golgi(GO:0033578) |
| 0.1 |
0.3 |
GO:0010761 |
fibroblast migration(GO:0010761) |
| 0.1 |
2.3 |
GO:0030866 |
cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 |
0.6 |
GO:0030903 |
notochord development(GO:0030903) |
| 0.1 |
1.2 |
GO:0002011 |
morphogenesis of an epithelial sheet(GO:0002011) |
| 0.1 |
0.3 |
GO:0034616 |
response to laminar fluid shear stress(GO:0034616) |
| 0.1 |
0.2 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 |
1.3 |
GO:0032964 |
collagen biosynthetic process(GO:0032964) |
| 0.1 |
0.3 |
GO:0032007 |
negative regulation of TOR signaling(GO:0032007) |
| 0.1 |
0.6 |
GO:0006379 |
mRNA cleavage(GO:0006379) |
| 0.1 |
0.3 |
GO:0019046 |
release from viral latency(GO:0019046) |
| 0.1 |
1.0 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 |
0.3 |
GO:0000394 |
RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 |
0.3 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 |
0.2 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.1 |
0.3 |
GO:0070542 |
response to fatty acid(GO:0070542) |
| 0.1 |
0.4 |
GO:0050666 |
regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.1 |
0.1 |
GO:0060684 |
epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.1 |
0.2 |
GO:0042536 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.1 |
1.0 |
GO:0048207 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.1 |
0.3 |
GO:0046349 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.1 |
0.3 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 |
0.2 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 |
0.2 |
GO:0032927 |
positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 |
0.5 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
0.5 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 |
0.2 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.1 |
0.2 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 |
0.5 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.0 |
0.1 |
GO:0006015 |
5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 |
0.1 |
GO:0006307 |
DNA dealkylation involved in DNA repair(GO:0006307) DNA dealkylation(GO:0035510) |
| 0.0 |
0.1 |
GO:0070407 |
oxidation-dependent protein catabolic process(GO:0070407) |
| 0.0 |
0.1 |
GO:0014887 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 |
0.2 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |
| 0.0 |
0.0 |
GO:0051788 |
response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 |
0.2 |
GO:0030037 |
actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.0 |
0.5 |
GO:0010875 |
positive regulation of cholesterol efflux(GO:0010875) |
| 0.0 |
1.0 |
GO:0042749 |
regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 |
0.0 |
GO:0021859 |
pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 |
0.1 |
GO:0090047 |
obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 |
0.7 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.2 |
GO:0060423 |
foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of branching involved in lung morphogenesis(GO:0061047) iris morphogenesis(GO:0061072) |
| 0.0 |
0.1 |
GO:0045992 |
negative regulation of embryonic development(GO:0045992) |
| 0.0 |
0.1 |
GO:1902807 |
negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 |
0.4 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.0 |
0.1 |
GO:0071941 |
nitrogen cycle metabolic process(GO:0071941) |
| 0.0 |
1.1 |
GO:0007398 |
ectoderm development(GO:0007398) |
| 0.0 |
0.6 |
GO:0000090 |
mitotic anaphase(GO:0000090) |
| 0.0 |
0.3 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.1 |
GO:0048668 |
hindbrain radial glia guided cell migration(GO:0021932) collateral sprouting(GO:0048668) regulation of collateral sprouting(GO:0048670) |
| 0.0 |
0.4 |
GO:0032092 |
positive regulation of protein binding(GO:0032092) |
| 0.0 |
2.2 |
GO:0046324 |
regulation of glucose import(GO:0046324) |
| 0.0 |
0.1 |
GO:0061188 |
regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 |
0.0 |
GO:1902803 |
regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 |
0.2 |
GO:0070669 |
response to interleukin-2(GO:0070669) |
| 0.0 |
0.2 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.0 |
1.3 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.0 |
0.5 |
GO:0001782 |
B cell homeostasis(GO:0001782) |
| 0.0 |
0.1 |
GO:0009301 |
snRNA transcription(GO:0009301) |
| 0.0 |
0.6 |
GO:0042346 |
positive regulation of NF-kappaB import into nucleus(GO:0042346) |
| 0.0 |
0.2 |
GO:0008054 |
negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 |
0.3 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
1.7 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.0 |
0.1 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 |
0.4 |
GO:0045652 |
regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 |
0.2 |
GO:0045778 |
positive regulation of ossification(GO:0045778) |
| 0.0 |
0.4 |
GO:0006743 |
ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.5 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.0 |
0.2 |
GO:0043392 |
negative regulation of DNA binding(GO:0043392) |
| 0.0 |
0.1 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 |
0.2 |
GO:0000098 |
sulfur amino acid catabolic process(GO:0000098) |
| 0.0 |
0.1 |
GO:0008298 |
intracellular mRNA localization(GO:0008298) |
| 0.0 |
0.1 |
GO:0009994 |
oocyte differentiation(GO:0009994) oocyte development(GO:0048599) |
| 0.0 |
0.1 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.0 |
1.7 |
GO:0071260 |
cellular response to mechanical stimulus(GO:0071260) |
| 0.0 |
0.1 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.0 |
0.9 |
GO:0007045 |
cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 |
0.0 |
GO:0032878 |
regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 |
0.5 |
GO:0050812 |
acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 |
0.2 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 |
0.0 |
GO:0031657 |
regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 |
0.1 |
GO:0051124 |
synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 |
0.1 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.0 |
0.1 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.3 |
GO:0034372 |
triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 |
0.1 |
GO:0090598 |
male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 |
0.4 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.0 |
0.1 |
GO:0042347 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) regulation of NF-kappaB import into nucleus(GO:0042345) negative regulation of NF-kappaB import into nucleus(GO:0042347) NF-kappaB import into nucleus(GO:0042348) |
| 0.0 |
0.1 |
GO:0006382 |
adenosine to inosine editing(GO:0006382) |
| 0.0 |
0.2 |
GO:0019388 |
galactose catabolic process(GO:0019388) |
| 0.0 |
0.0 |
GO:0032707 |
negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 |
0.2 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.1 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.0 |
0.1 |
GO:0045945 |
positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 |
0.1 |
GO:0099515 |
nuclear migration along microfilament(GO:0031022) actin filament-based transport(GO:0099515) |
| 0.0 |
0.3 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 |
0.1 |
GO:0046292 |
formaldehyde metabolic process(GO:0046292) |
| 0.0 |
0.1 |
GO:1903301 |
regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 |
0.1 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 |
0.2 |
GO:0050858 |
negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.2 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 |
0.6 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.0 |
0.3 |
GO:0006308 |
DNA catabolic process(GO:0006308) |
| 0.0 |
0.1 |
GO:0007095 |
mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 |
0.1 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.0 |
0.2 |
GO:0007435 |
salivary gland morphogenesis(GO:0007435) |
| 0.0 |
1.1 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.0 |
0.7 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
0.4 |
GO:0006972 |
hyperosmotic response(GO:0006972) |
| 0.0 |
0.1 |
GO:0090343 |
positive regulation of cell aging(GO:0090343) |
| 0.0 |
0.2 |
GO:0050872 |
white fat cell differentiation(GO:0050872) |
| 0.0 |
0.2 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
0.0 |
GO:0032196 |
transposition, DNA-mediated(GO:0006313) transposition(GO:0032196) |
| 0.0 |
0.1 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.0 |
0.3 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.0 |
0.2 |
GO:0006553 |
lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 |
0.1 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.3 |
GO:0034394 |
protein localization to cell surface(GO:0034394) |
| 0.0 |
0.1 |
GO:0000012 |
single strand break repair(GO:0000012) |
| 0.0 |
0.6 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.1 |
GO:0071542 |
dopaminergic neuron differentiation(GO:0071542) |
| 0.0 |
0.2 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.0 |
0.1 |
GO:0030917 |
midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 |
0.1 |
GO:0016264 |
gap junction assembly(GO:0016264) |
| 0.0 |
0.2 |
GO:0001961 |
positive regulation of cytokine-mediated signaling pathway(GO:0001961) |
| 0.0 |
0.1 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) negative regulation of sodium ion transport(GO:0010766) |
| 0.0 |
0.1 |
GO:0010955 |
negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 |
0.0 |
GO:0009948 |
anterior/posterior axis specification(GO:0009948) |
| 0.0 |
0.1 |
GO:0090083 |
inclusion body assembly(GO:0070841) regulation of inclusion body assembly(GO:0090083) |
| 0.0 |
0.3 |
GO:0008347 |
glial cell migration(GO:0008347) |
| 0.0 |
0.2 |
GO:0043954 |
cell junction maintenance(GO:0034331) cellular component maintenance(GO:0043954) cell-cell junction maintenance(GO:0045217) |
| 0.0 |
0.1 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.0 |
0.0 |
GO:1903519 |
apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) regulation of apoptotic process involved in morphogenesis(GO:1902337) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) regulation of apoptotic process involved in development(GO:1904748) |
| 0.0 |
0.2 |
GO:0006910 |
phagocytosis, recognition(GO:0006910) |
| 0.0 |
0.4 |
GO:0010463 |
mesenchymal cell proliferation(GO:0010463) |
| 0.0 |
0.1 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 |
0.1 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.0 |
0.7 |
GO:0060041 |
retina development in camera-type eye(GO:0060041) |
| 0.0 |
0.1 |
GO:0001881 |
receptor recycling(GO:0001881) |
| 0.0 |
0.1 |
GO:0050910 |
detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 |
0.7 |
GO:0007602 |
phototransduction(GO:0007602) |
| 0.0 |
1.2 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
0.1 |
GO:0071364 |
cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 |
0.1 |
GO:2000398 |
regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 |
0.2 |
GO:0006884 |
cell volume homeostasis(GO:0006884) |
| 0.0 |
0.2 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.0 |
0.1 |
GO:0045165 |
cell fate commitment(GO:0045165) |
| 0.0 |
0.1 |
GO:0031573 |
intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 |
0.3 |
GO:0050892 |
intestinal absorption(GO:0050892) |
| 0.0 |
0.1 |
GO:0006290 |
pyrimidine dimer repair(GO:0006290) |
| 0.0 |
0.1 |
GO:0048486 |
parasympathetic nervous system development(GO:0048486) |
| 0.0 |
0.1 |
GO:0003084 |
positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 |
0.1 |
GO:0035246 |
peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 |
0.1 |
GO:0060828 |
regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 |
0.2 |
GO:0007176 |
regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 |
1.4 |
GO:0008154 |
actin polymerization or depolymerization(GO:0008154) |
| 0.0 |
0.1 |
GO:0045329 |
amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 |
0.1 |
GO:0009414 |
response to water deprivation(GO:0009414) response to water(GO:0009415) paranodal junction assembly(GO:0030913) |
| 0.0 |
0.1 |
GO:0006069 |
ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 |
0.5 |
GO:0045669 |
positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 |
0.1 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.0 |
0.1 |
GO:2000095 |
regulation of non-canonical Wnt signaling pathway(GO:2000050) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 |
0.2 |
GO:0043114 |
regulation of vascular permeability(GO:0043114) |
| 0.0 |
0.1 |
GO:0009642 |
response to light intensity(GO:0009642) |
| 0.0 |
0.1 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.0 |
0.3 |
GO:0051897 |
positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 |
0.1 |
GO:0043923 |
positive regulation by host of viral transcription(GO:0043923) |
| 0.0 |
0.2 |
GO:0060538 |
skeletal muscle organ development(GO:0060538) |
| 0.0 |
0.0 |
GO:0045065 |
cytotoxic T cell differentiation(GO:0045065) |
| 0.0 |
0.2 |
GO:0045161 |
neuronal ion channel clustering(GO:0045161) |
| 0.0 |
0.2 |
GO:0042036 |
negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 |
0.1 |
GO:0006084 |
acetyl-CoA metabolic process(GO:0006084) |
| 0.0 |
0.2 |
GO:0043525 |
positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 |
0.1 |
GO:0019321 |
pentose metabolic process(GO:0019321) |
| 0.0 |
0.1 |
GO:0071514 |
genetic imprinting(GO:0071514) |
| 0.0 |
0.2 |
GO:0008214 |
protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 |
0.9 |
GO:0007588 |
excretion(GO:0007588) |
| 0.0 |
0.1 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.0 |
0.0 |
GO:0052510 |
induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 |
0.2 |
GO:0006891 |
intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 |
0.2 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.0 |
GO:0048549 |
regulation of pinocytosis(GO:0048548) positive regulation of pinocytosis(GO:0048549) |
| 0.0 |
0.1 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.0 |
0.9 |
GO:0019882 |
antigen processing and presentation(GO:0019882) |
| 0.0 |
0.2 |
GO:0042752 |
regulation of circadian rhythm(GO:0042752) |
| 0.0 |
0.0 |
GO:0007538 |
primary sex determination(GO:0007538) |
| 0.0 |
0.0 |
GO:0070120 |
ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 |
0.3 |
GO:0045768 |
obsolete positive regulation of anti-apoptosis(GO:0045768) |
| 0.0 |
0.3 |
GO:0016236 |
macroautophagy(GO:0016236) |
| 0.0 |
0.3 |
GO:0032088 |
negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 |
0.1 |
GO:0006098 |
pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |