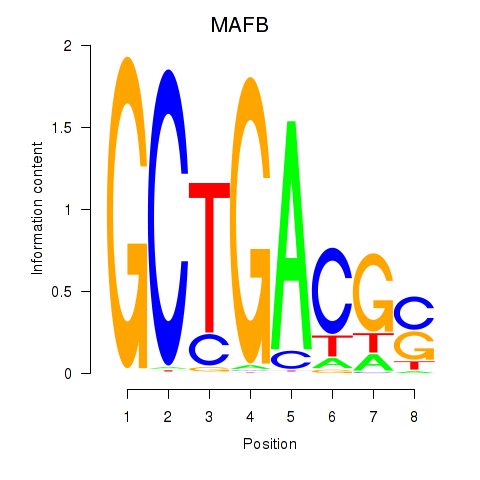
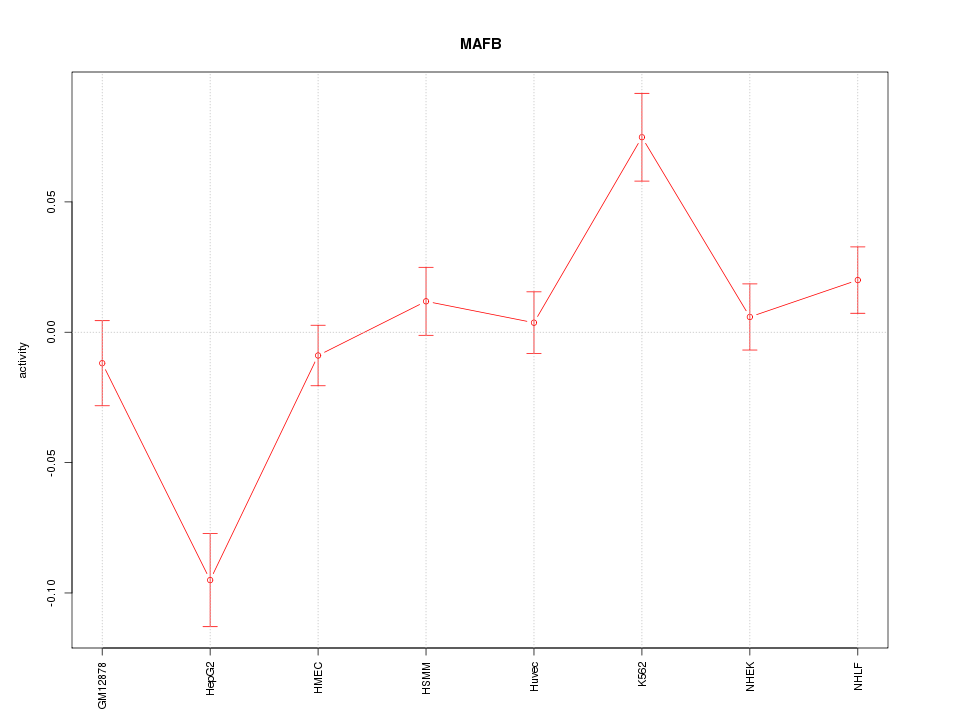
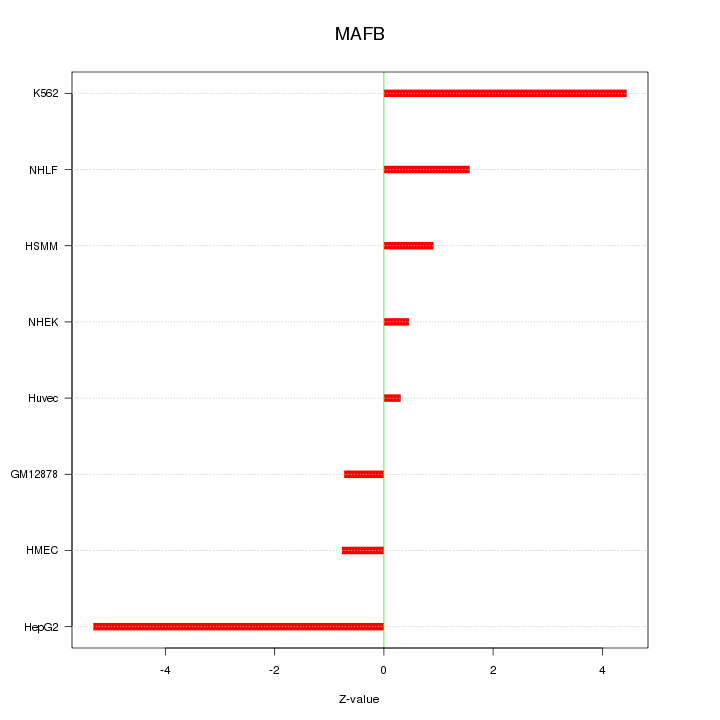
| 1.4 |
4.2 |
GO:0030091 |
protein repair(GO:0030091) |
| 1.0 |
3.0 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.8 |
4.1 |
GO:0030210 |
heparin biosynthetic process(GO:0030210) negative regulation of vascular permeability(GO:0043116) Tie signaling pathway(GO:0048014) |
| 0.6 |
1.9 |
GO:0035408 |
histone H3-T6 phosphorylation(GO:0035408) |
| 0.5 |
1.6 |
GO:0051083 |
'de novo' cotranslational protein folding(GO:0051083) |
| 0.5 |
3.7 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.5 |
2.5 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.5 |
1.0 |
GO:0060599 |
lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.5 |
1.4 |
GO:0006549 |
isoleucine metabolic process(GO:0006549) |
| 0.4 |
1.6 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 0.4 |
1.2 |
GO:0051255 |
spindle midzone assembly(GO:0051255) meiotic spindle midzone assembly(GO:0051257) |
| 0.4 |
1.9 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.4 |
0.8 |
GO:0070841 |
inclusion body assembly(GO:0070841) |
| 0.4 |
1.1 |
GO:0046586 |
regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.4 |
1.1 |
GO:0090047 |
obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.3 |
2.4 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.3 |
0.9 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.3 |
1.6 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 |
0.9 |
GO:0034625 |
fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.3 |
0.9 |
GO:0010041 |
response to iron(III) ion(GO:0010041) positive regulation of histone phosphorylation(GO:0033129) response to DDT(GO:0046680) regulation of chromosome condensation(GO:0060623) cellular response to iron ion(GO:0071281) cellular response to iron(III) ion(GO:0071283) |
| 0.3 |
1.5 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.3 |
1.5 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 |
1.4 |
GO:0032959 |
inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.3 |
2.0 |
GO:0035058 |
nonmotile primary cilium assembly(GO:0035058) |
| 0.3 |
2.3 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.3 |
3.7 |
GO:0031498 |
nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.3 |
2.0 |
GO:0030007 |
cellular potassium ion homeostasis(GO:0030007) negative regulation of cell volume(GO:0045794) |
| 0.3 |
1.1 |
GO:0000389 |
mRNA 3'-splice site recognition(GO:0000389) |
| 0.3 |
3.0 |
GO:0000076 |
DNA replication checkpoint(GO:0000076) |
| 0.3 |
0.8 |
GO:0048311 |
mitochondrion distribution(GO:0048311) |
| 0.3 |
0.8 |
GO:0042501 |
serine phosphorylation of STAT protein(GO:0042501) |
| 0.3 |
1.6 |
GO:0015742 |
alpha-ketoglutarate transport(GO:0015742) |
| 0.3 |
1.8 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.3 |
1.0 |
GO:0009956 |
radial pattern formation(GO:0009956) |
| 0.3 |
1.0 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.3 |
1.3 |
GO:0017085 |
response to insecticide(GO:0017085) |
| 0.3 |
0.8 |
GO:0097237 |
cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.2 |
0.7 |
GO:0046604 |
regulation of mitotic centrosome separation(GO:0046602) positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 |
0.7 |
GO:2000644 |
negative regulation of low-density lipoprotein particle clearance(GO:0010989) low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.2 |
1.2 |
GO:0032933 |
SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 |
1.1 |
GO:0060528 |
secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.2 |
0.7 |
GO:2000105 |
positive regulation of DNA-dependent DNA replication initiation(GO:0032298) positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.2 |
0.7 |
GO:0014724 |
regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) mitochondrial calcium ion homeostasis(GO:0051560) positive regulation of mitochondrial calcium ion concentration(GO:0051561) relaxation of skeletal muscle(GO:0090076) |
| 0.2 |
1.5 |
GO:0050832 |
response to fungus(GO:0009620) defense response to fungus(GO:0050832) |
| 0.2 |
1.1 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.2 |
2.3 |
GO:0009263 |
deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.2 |
0.6 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.2 |
0.8 |
GO:0071863 |
cell proliferation in bone marrow(GO:0071838) apoptotic process in bone marrow(GO:0071839) regulation of cell proliferation in bone marrow(GO:0071863) positive regulation of cell proliferation in bone marrow(GO:0071864) regulation of apoptotic process in bone marrow(GO:0071865) negative regulation of apoptotic process in bone marrow(GO:0071866) |
| 0.2 |
3.8 |
GO:0060765 |
regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.2 |
0.8 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.2 |
0.4 |
GO:0030952 |
establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.2 |
1.8 |
GO:0006474 |
N-terminal protein amino acid acetylation(GO:0006474) |
| 0.2 |
1.0 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.2 |
0.9 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 |
1.1 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.2 |
0.7 |
GO:0006391 |
transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 |
0.5 |
GO:0006740 |
NADPH regeneration(GO:0006740) |
| 0.2 |
0.9 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.2 |
0.5 |
GO:1904036 |
negative regulation of epithelial cell apoptotic process(GO:1904036) negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.2 |
0.2 |
GO:0031018 |
endocrine pancreas development(GO:0031018) |
| 0.2 |
0.5 |
GO:0034036 |
purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 |
0.5 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.2 |
0.5 |
GO:0034770 |
histone H4-K20 methylation(GO:0034770) |
| 0.2 |
0.9 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.2 |
1.0 |
GO:0001514 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 |
0.7 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.2 |
0.5 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 |
0.5 |
GO:0019918 |
peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.2 |
0.3 |
GO:0097154 |
cerebral cortex GABAergic interneuron differentiation(GO:0021892) GABAergic neuron differentiation(GO:0097154) |
| 0.2 |
0.8 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.2 |
0.9 |
GO:0043316 |
natural killer cell activation involved in immune response(GO:0002323) cytotoxic T cell degranulation(GO:0043316) natural killer cell degranulation(GO:0043320) |
| 0.2 |
0.3 |
GO:0034371 |
chylomicron remodeling(GO:0034371) |
| 0.2 |
0.9 |
GO:0051051 |
negative regulation of transport(GO:0051051) |
| 0.2 |
0.6 |
GO:0045199 |
maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 |
0.2 |
GO:0003149 |
membranous septum morphogenesis(GO:0003149) |
| 0.2 |
1.2 |
GO:0030422 |
production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 |
1.0 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
0.3 |
GO:0060285 |
cilium-dependent cell motility(GO:0060285) |
| 0.1 |
0.4 |
GO:0006591 |
ornithine metabolic process(GO:0006591) |
| 0.1 |
0.4 |
GO:0016926 |
protein desumoylation(GO:0016926) |
| 0.1 |
0.1 |
GO:0048755 |
branching morphogenesis of a nerve(GO:0048755) |
| 0.1 |
1.3 |
GO:0006047 |
UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 |
1.8 |
GO:0048261 |
negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.1 |
0.4 |
GO:0006307 |
DNA dealkylation involved in DNA repair(GO:0006307) DNA dealkylation(GO:0035510) |
| 0.1 |
2.0 |
GO:0006787 |
porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 |
2.6 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 |
0.4 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.1 |
3.6 |
GO:0006911 |
phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.1 |
0.9 |
GO:0045010 |
actin nucleation(GO:0045010) |
| 0.1 |
1.5 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
1.7 |
GO:0032331 |
negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 |
0.7 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 |
0.4 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.1 |
0.8 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 |
0.6 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 |
0.4 |
GO:0032914 |
positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 |
0.3 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 |
0.6 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 |
0.5 |
GO:0045900 |
negative regulation of translational elongation(GO:0045900) |
| 0.1 |
0.4 |
GO:0035470 |
positive regulation of vascular wound healing(GO:0035470) |
| 0.1 |
0.5 |
GO:0018352 |
protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 |
0.4 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 |
0.4 |
GO:0045062 |
extrathymic T cell differentiation(GO:0033078) extrathymic T cell selection(GO:0045062) |
| 0.1 |
0.4 |
GO:0009996 |
negative regulation of cell fate specification(GO:0009996) positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 |
0.8 |
GO:0021830 |
interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.1 |
1.1 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.1 |
0.2 |
GO:0072033 |
renal vesicle formation(GO:0072033) |
| 0.1 |
1.0 |
GO:0045008 |
depyrimidination(GO:0045008) |
| 0.1 |
0.4 |
GO:0001807 |
type IV hypersensitivity(GO:0001806) regulation of type IV hypersensitivity(GO:0001807) |
| 0.1 |
0.3 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 |
0.5 |
GO:0070542 |
response to fatty acid(GO:0070542) |
| 0.1 |
0.8 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.1 |
0.3 |
GO:0033630 |
positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.1 |
1.1 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 |
1.0 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 |
0.3 |
GO:0048549 |
positive regulation of pinocytosis(GO:0048549) |
| 0.1 |
0.3 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 |
0.1 |
GO:1903514 |
regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.1 |
0.6 |
GO:0010957 |
negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 |
3.0 |
GO:0043496 |
regulation of protein homodimerization activity(GO:0043496) |
| 0.1 |
0.3 |
GO:0032581 |
ER-dependent peroxisome organization(GO:0032581) |
| 0.1 |
0.9 |
GO:0034755 |
iron ion transmembrane transport(GO:0034755) |
| 0.1 |
1.0 |
GO:0071692 |
sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 |
0.5 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.1 |
1.1 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 |
0.9 |
GO:0022601 |
menstrual cycle phase(GO:0022601) |
| 0.1 |
0.6 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 |
0.7 |
GO:0016559 |
peroxisome fission(GO:0016559) |
| 0.1 |
0.3 |
GO:0001837 |
epithelial to mesenchymal transition(GO:0001837) |
| 0.1 |
1.0 |
GO:0019359 |
nicotinamide nucleotide biosynthetic process(GO:0019359) |
| 0.1 |
0.3 |
GO:0009953 |
dorsal/ventral pattern formation(GO:0009953) |
| 0.1 |
0.4 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.1 |
1.6 |
GO:0032332 |
positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 |
0.8 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.1 |
0.2 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.1 |
0.4 |
GO:0006498 |
N-terminal protein lipidation(GO:0006498) |
| 0.1 |
0.2 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.1 |
0.9 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 |
0.6 |
GO:0032527 |
retrograde protein transport, ER to cytosol(GO:0030970) protein exit from endoplasmic reticulum(GO:0032527) |
| 0.1 |
0.2 |
GO:0060557 |
positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 |
0.7 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.1 |
0.7 |
GO:0090400 |
stress-induced premature senescence(GO:0090400) |
| 0.1 |
1.0 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.1 |
1.4 |
GO:0018298 |
protein-chromophore linkage(GO:0018298) |
| 0.1 |
0.4 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
2.4 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |
| 0.1 |
2.1 |
GO:0032781 |
positive regulation of ATPase activity(GO:0032781) |
| 0.1 |
0.3 |
GO:0060012 |
synaptic transmission, glycinergic(GO:0060012) |
| 0.1 |
0.2 |
GO:0033091 |
positive regulation of immature T cell proliferation(GO:0033091) |
| 0.1 |
0.6 |
GO:0006196 |
AMP catabolic process(GO:0006196) |
| 0.1 |
0.3 |
GO:0007109 |
obsolete cytokinesis, completion of separation(GO:0007109) |
| 0.1 |
0.4 |
GO:0046022 |
regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 |
0.6 |
GO:0006346 |
methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 |
0.2 |
GO:0007619 |
courtship behavior(GO:0007619) male courtship behavior(GO:0008049) male mating behavior(GO:0060179) |
| 0.1 |
0.4 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.1 |
0.3 |
GO:0007500 |
mesodermal cell fate determination(GO:0007500) |
| 0.1 |
0.7 |
GO:0071786 |
endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 |
0.2 |
GO:0032899 |
regulation of neurotrophin production(GO:0032899) negative regulation of neurotrophin production(GO:0032900) |
| 0.1 |
2.5 |
GO:0070534 |
protein K63-linked ubiquitination(GO:0070534) |
| 0.1 |
0.2 |
GO:0045080 |
positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.1 |
0.3 |
GO:2001258 |
negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) negative regulation of calcium ion transmembrane transporter activity(GO:1901020) negative regulation of cation channel activity(GO:2001258) |
| 0.1 |
0.5 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.1 |
0.9 |
GO:0033523 |
histone monoubiquitination(GO:0010390) histone H2B ubiquitination(GO:0033523) |
| 0.1 |
0.1 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.1 |
0.7 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.1 |
0.7 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 |
0.7 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.1 |
2.2 |
GO:0006309 |
DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.1 |
1.9 |
GO:0000722 |
telomere maintenance via recombination(GO:0000722) |
| 0.1 |
0.1 |
GO:0045908 |
negative regulation of vasodilation(GO:0045908) |
| 0.1 |
0.5 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 |
0.5 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.1 |
0.2 |
GO:0006344 |
maintenance of chromatin silencing(GO:0006344) hair follicle placode formation(GO:0060789) fungiform papilla development(GO:0061196) fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 0.1 |
0.2 |
GO:0003128 |
heart field specification(GO:0003128) heart induction(GO:0003129) |
| 0.1 |
0.2 |
GO:0001973 |
adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 |
0.2 |
GO:0070781 |
response to biotin(GO:0070781) |
| 0.1 |
0.2 |
GO:0031034 |
skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 |
1.5 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.1 |
0.6 |
GO:0006072 |
glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 |
0.4 |
GO:0045109 |
intermediate filament organization(GO:0045109) |
| 0.1 |
1.1 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.1 |
0.4 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.1 |
0.4 |
GO:0090136 |
epithelial cell-cell adhesion(GO:0090136) |
| 0.1 |
0.4 |
GO:0001967 |
suckling behavior(GO:0001967) equilibrioception(GO:0050957) |
| 0.1 |
0.3 |
GO:0006642 |
triglyceride mobilization(GO:0006642) |
| 0.1 |
0.1 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 |
0.3 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 |
0.9 |
GO:0034405 |
response to fluid shear stress(GO:0034405) |
| 0.1 |
0.3 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 |
1.0 |
GO:0000090 |
mitotic anaphase(GO:0000090) |
| 0.1 |
0.5 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.1 |
0.3 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.1 |
0.3 |
GO:0071436 |
sodium ion export(GO:0071436) |
| 0.1 |
0.8 |
GO:0001921 |
positive regulation of receptor recycling(GO:0001921) |
| 0.1 |
0.3 |
GO:0043524 |
negative regulation of neuron apoptotic process(GO:0043524) negative regulation of neuron death(GO:1901215) |
| 0.1 |
0.6 |
GO:0048892 |
sensory system development(GO:0048880) lateral line nerve development(GO:0048892) lateral line nerve glial cell differentiation(GO:0048895) lateral line system development(GO:0048925) lateral line nerve glial cell development(GO:0048937) iridophore differentiation(GO:0050935) |
| 0.1 |
0.2 |
GO:0006542 |
glutamine biosynthetic process(GO:0006542) |
| 0.1 |
0.2 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.1 |
0.1 |
GO:0015870 |
acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.1 |
0.2 |
GO:0008637 |
apoptotic mitochondrial changes(GO:0008637) |
| 0.1 |
0.2 |
GO:0045003 |
DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.1 |
0.2 |
GO:0045898 |
regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 |
1.5 |
GO:0044349 |
nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) |
| 0.1 |
0.3 |
GO:0019987 |
obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.1 |
0.4 |
GO:0001890 |
placenta development(GO:0001890) |
| 0.1 |
0.1 |
GO:0060744 |
thelarche(GO:0042695) development of secondary sexual characteristics(GO:0045136) development of secondary female sexual characteristics(GO:0046543) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 |
0.2 |
GO:0046831 |
regulation of RNA export from nucleus(GO:0046831) |
| 0.1 |
0.4 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.1 |
0.1 |
GO:0060896 |
neural plate anterior/posterior regionalization(GO:0021999) neural plate pattern specification(GO:0060896) neural plate regionalization(GO:0060897) |
| 0.1 |
2.0 |
GO:0033572 |
ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 |
0.2 |
GO:0061364 |
response to cortisol(GO:0051414) apoptotic process involved in luteolysis(GO:0061364) |
| 0.1 |
0.2 |
GO:0021801 |
cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.1 |
1.9 |
GO:0006220 |
pyrimidine nucleotide metabolic process(GO:0006220) |
| 0.1 |
0.3 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.1 |
0.1 |
GO:0010661 |
positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 |
0.6 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 |
0.1 |
GO:1904396 |
regulation of synaptic growth at neuromuscular junction(GO:0008582) regulation of neuromuscular junction development(GO:1904396) |
| 0.1 |
0.2 |
GO:0061084 |
regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) negative regulation of protein folding(GO:1903333) |
| 0.1 |
3.1 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.1 |
0.1 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 |
0.9 |
GO:1901661 |
quinone metabolic process(GO:1901661) |
| 0.0 |
0.1 |
GO:0032230 |
positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.0 |
0.2 |
GO:0015917 |
aminophospholipid transport(GO:0015917) |
| 0.0 |
0.2 |
GO:0009133 |
nucleoside diphosphate biosynthetic process(GO:0009133) |
| 0.0 |
0.3 |
GO:0051602 |
response to electrical stimulus(GO:0051602) |
| 0.0 |
0.1 |
GO:0060370 |
susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 |
0.2 |
GO:0021937 |
cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 |
0.1 |
GO:0018243 |
protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 |
0.8 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
0.2 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
0.5 |
GO:0001945 |
lymph vessel development(GO:0001945) |
| 0.0 |
0.5 |
GO:0048169 |
regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 |
0.2 |
GO:0021955 |
central nervous system projection neuron axonogenesis(GO:0021952) central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 |
0.9 |
GO:0043278 |
response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 |
0.5 |
GO:0007007 |
inner mitochondrial membrane organization(GO:0007007) |
| 0.0 |
0.7 |
GO:1903078 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 |
0.8 |
GO:0051703 |
social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 |
0.2 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.0 |
2.3 |
GO:0050434 |
positive regulation of viral transcription(GO:0050434) |
| 0.0 |
0.3 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.1 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.0 |
0.1 |
GO:0050917 |
sensory perception of umami taste(GO:0050917) |
| 0.0 |
0.4 |
GO:0045542 |
positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 |
0.2 |
GO:0035020 |
regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 |
0.4 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 |
0.1 |
GO:0015959 |
diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 |
0.8 |
GO:0050812 |
acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 |
0.8 |
GO:0000380 |
alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 |
0.8 |
GO:0006596 |
polyamine biosynthetic process(GO:0006596) |
| 0.0 |
0.3 |
GO:0060314 |
regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 |
0.5 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 |
0.9 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.0 |
0.2 |
GO:0007635 |
chemosensory behavior(GO:0007635) |
| 0.0 |
0.5 |
GO:0032784 |
regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 |
0.7 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.1 |
GO:0061030 |
axon target recognition(GO:0007412) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 |
0.3 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.0 |
0.2 |
GO:0007468 |
regulation of rhodopsin gene expression(GO:0007468) retinal metabolic process(GO:0042574) |
| 0.0 |
0.5 |
GO:0007223 |
Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 |
0.1 |
GO:0070092 |
regulation of transmission of nerve impulse(GO:0051969) positive regulation of transmission of nerve impulse(GO:0051971) regulation of glucagon secretion(GO:0070092) somatostatin secretion(GO:0070253) |
| 0.0 |
1.1 |
GO:0006626 |
protein targeting to mitochondrion(GO:0006626) |
| 0.0 |
0.1 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.0 |
0.1 |
GO:0010761 |
fibroblast migration(GO:0010761) |
| 0.0 |
0.3 |
GO:0031427 |
response to methotrexate(GO:0031427) |
| 0.0 |
0.2 |
GO:2000179 |
positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 |
0.3 |
GO:0051017 |
actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 |
0.7 |
GO:0046676 |
negative regulation of insulin secretion(GO:0046676) |
| 0.0 |
0.7 |
GO:0060393 |
regulation of pathway-restricted SMAD protein phosphorylation(GO:0060393) |
| 0.0 |
0.2 |
GO:0060421 |
positive regulation of cardiac muscle tissue growth(GO:0055023) positive regulation of cardiac muscle tissue development(GO:0055025) positive regulation of cardiac muscle cell proliferation(GO:0060045) positive regulation of heart growth(GO:0060421) |
| 0.0 |
1.1 |
GO:0045980 |
negative regulation of nucleotide metabolic process(GO:0045980) |
| 0.0 |
0.1 |
GO:0048537 |
mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 |
0.5 |
GO:0097031 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.2 |
GO:0021520 |
spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 |
2.9 |
GO:0006521 |
regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 |
0.1 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.0 |
0.1 |
GO:0006122 |
mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 |
0.5 |
GO:0060638 |
mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 |
0.8 |
GO:0007172 |
signal complex assembly(GO:0007172) |
| 0.0 |
0.3 |
GO:0051963 |
regulation of synapse assembly(GO:0051963) |
| 0.0 |
0.1 |
GO:0006379 |
mRNA cleavage(GO:0006379) RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.0 |
0.1 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 |
0.3 |
GO:0045059 |
positive thymic T cell selection(GO:0045059) |
| 0.0 |
0.1 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 |
0.0 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 |
2.2 |
GO:0006892 |
post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 |
0.2 |
GO:0033363 |
secretory granule organization(GO:0033363) |
| 0.0 |
0.3 |
GO:0042517 |
positive regulation of tyrosine phosphorylation of Stat3 protein(GO:0042517) |
| 0.0 |
0.5 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.0 |
0.3 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.1 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.7 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.9 |
GO:0006413 |
translational initiation(GO:0006413) |
| 0.0 |
0.2 |
GO:0030578 |
nuclear body organization(GO:0030575) PML body organization(GO:0030578) |
| 0.0 |
4.1 |
GO:0006417 |
regulation of translation(GO:0006417) |
| 0.0 |
0.1 |
GO:0006924 |
activation-induced cell death of T cells(GO:0006924) |
| 0.0 |
0.1 |
GO:0010544 |
negative regulation of platelet activation(GO:0010544) |
| 0.0 |
0.5 |
GO:1901799 |
negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) negative regulation of proteasomal protein catabolic process(GO:1901799) negative regulation of proteolysis involved in cellular protein catabolic process(GO:1903051) negative regulation of cellular protein catabolic process(GO:1903363) |
| 0.0 |
0.1 |
GO:0002606 |
dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 |
0.8 |
GO:0001656 |
metanephros development(GO:0001656) |
| 0.0 |
0.1 |
GO:0008356 |
asymmetric cell division(GO:0008356) |
| 0.0 |
0.1 |
GO:0009438 |
methylglyoxal metabolic process(GO:0009438) |
| 0.0 |
0.2 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.0 |
0.6 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.1 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.0 |
0.1 |
GO:0032402 |
establishment of melanosome localization(GO:0032401) melanosome transport(GO:0032402) pigment granule transport(GO:0051904) establishment of pigment granule localization(GO:0051905) |
| 0.0 |
0.3 |
GO:0017121 |
phospholipid scrambling(GO:0017121) |
| 0.0 |
0.2 |
GO:0043462 |
regulation of ATPase activity(GO:0043462) |
| 0.0 |
0.3 |
GO:0060674 |
placenta blood vessel development(GO:0060674) |
| 0.0 |
0.3 |
GO:0032703 |
negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 |
0.6 |
GO:0031055 |
DNA replication-independent nucleosome assembly(GO:0006336) chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) DNA replication-independent nucleosome organization(GO:0034724) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 |
0.1 |
GO:0032097 |
positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) positive regulation of response to extracellular stimulus(GO:0032106) positive regulation of response to nutrient levels(GO:0032109) |
| 0.0 |
0.1 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.3 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 |
0.1 |
GO:0033211 |
adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 |
0.1 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 |
0.1 |
GO:0060571 |
morphogenesis of an epithelial fold(GO:0060571) |
| 0.0 |
0.3 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.0 |
0.3 |
GO:0045329 |
amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 |
0.1 |
GO:0035455 |
response to interferon-alpha(GO:0035455) |
| 0.0 |
0.2 |
GO:0007080 |
mitotic metaphase plate congression(GO:0007080) |
| 0.0 |
0.2 |
GO:0033235 |
regulation of protein sumoylation(GO:0033233) positive regulation of protein sumoylation(GO:0033235) |
| 0.0 |
1.4 |
GO:0051084 |
'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 |
2.7 |
GO:0006364 |
rRNA processing(GO:0006364) |
| 0.0 |
1.6 |
GO:0009408 |
response to heat(GO:0009408) |
| 0.0 |
0.1 |
GO:0001964 |
startle response(GO:0001964) |
| 0.0 |
0.7 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.2 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.2 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.0 |
0.1 |
GO:0036260 |
7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 |
0.1 |
GO:0032494 |
response to peptidoglycan(GO:0032494) |
| 0.0 |
0.1 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 0.0 |
0.3 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.0 |
1.6 |
GO:0061025 |
membrane fusion(GO:0061025) |
| 0.0 |
0.4 |
GO:0032233 |
positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 |
0.2 |
GO:0048485 |
sympathetic nervous system development(GO:0048485) |
| 0.0 |
0.4 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.5 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 |
0.1 |
GO:0042339 |
keratan sulfate metabolic process(GO:0042339) |
| 0.0 |
1.1 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.5 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.2 |
GO:0010799 |
regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 |
0.1 |
GO:0042516 |
tyrosine phosphorylation of Stat3 protein(GO:0042503) regulation of tyrosine phosphorylation of Stat3 protein(GO:0042516) |
| 0.0 |
0.0 |
GO:0051547 |
regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 |
0.2 |
GO:0016051 |
carbohydrate biosynthetic process(GO:0016051) |
| 0.0 |
0.5 |
GO:0000725 |
double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 |
0.1 |
GO:0048295 |
positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 |
0.1 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 |
0.2 |
GO:0031958 |
corticosteroid receptor signaling pathway(GO:0031958) glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 |
0.1 |
GO:0050758 |
thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 |
0.2 |
GO:0007143 |
female meiotic division(GO:0007143) |
| 0.0 |
0.2 |
GO:0019725 |
cellular homeostasis(GO:0019725) |
| 0.0 |
0.4 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.0 |
0.4 |
GO:0007140 |
male meiosis(GO:0007140) |
| 0.0 |
0.2 |
GO:0046655 |
folic acid metabolic process(GO:0046655) |
| 0.0 |
0.4 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.4 |
GO:0030032 |
lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 |
0.9 |
GO:0048738 |
cardiac muscle tissue development(GO:0048738) |
| 0.0 |
0.7 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.0 |
0.5 |
GO:0050690 |
regulation of defense response to virus by virus(GO:0050690) |
| 0.0 |
1.4 |
GO:0006402 |
mRNA catabolic process(GO:0006402) |
| 0.0 |
0.1 |
GO:0034765 |
regulation of ion transmembrane transport(GO:0034765) |
| 0.0 |
0.0 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
0.3 |
GO:0030641 |
regulation of cellular pH(GO:0030641) |
| 0.0 |
0.5 |
GO:0006182 |
cGMP biosynthetic process(GO:0006182) |
| 0.0 |
0.6 |
GO:0007052 |
mitotic spindle organization(GO:0007052) |
| 0.0 |
0.6 |
GO:0042254 |
ribosome biogenesis(GO:0042254) |
| 0.0 |
0.1 |
GO:0042727 |
flavin-containing compound metabolic process(GO:0042726) flavin-containing compound biosynthetic process(GO:0042727) |
| 0.0 |
0.4 |
GO:0000209 |
protein polyubiquitination(GO:0000209) |
| 0.0 |
0.2 |
GO:0015884 |
folic acid transport(GO:0015884) |
| 0.0 |
1.2 |
GO:0006367 |
transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 |
0.1 |
GO:0050854 |
regulation of antigen receptor-mediated signaling pathway(GO:0050854) regulation of T cell receptor signaling pathway(GO:0050856) negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.1 |
GO:0043615 |
astrocyte cell migration(GO:0043615) |
| 0.0 |
0.1 |
GO:0071357 |
response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 |
0.3 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.3 |
GO:0033138 |
positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 |
1.0 |
GO:0031497 |
chromatin assembly(GO:0031497) |
| 0.0 |
0.2 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.0 |
0.1 |
GO:0030917 |
midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 |
0.2 |
GO:0042246 |
tissue regeneration(GO:0042246) |
| 0.0 |
0.8 |
GO:0007126 |
meiotic nuclear division(GO:0007126) |
| 0.0 |
0.0 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.0 |
0.1 |
GO:0006469 |
negative regulation of protein kinase activity(GO:0006469) |
| 0.0 |
0.1 |
GO:0031122 |
cytoplasmic microtubule organization(GO:0031122) |
| 0.0 |
0.1 |
GO:0060742 |
epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 |
0.3 |
GO:0000768 |
syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 |
0.4 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.3 |
GO:0042776 |
mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 |
0.8 |
GO:0008033 |
tRNA processing(GO:0008033) |
| 0.0 |
0.5 |
GO:0031111 |
negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 |
0.2 |
GO:0045730 |
respiratory burst(GO:0045730) |
| 0.0 |
0.5 |
GO:0051149 |
positive regulation of muscle cell differentiation(GO:0051149) |
| 0.0 |
0.9 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
2.2 |
GO:0034329 |
cell junction assembly(GO:0034329) |
| 0.0 |
0.1 |
GO:0090231 |
regulation of spindle checkpoint(GO:0090231) |
| 0.0 |
1.6 |
GO:0006415 |
translational termination(GO:0006415) |
| 0.0 |
0.1 |
GO:0031290 |
retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 |
0.0 |
GO:0014829 |
tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) vein smooth muscle contraction(GO:0014826) vascular smooth muscle contraction(GO:0014829) |
| 0.0 |
0.1 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.2 |
GO:0007368 |
determination of left/right symmetry(GO:0007368) specification of symmetry(GO:0009799) determination of bilateral symmetry(GO:0009855) |
| 0.0 |
0.1 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.0 |
0.3 |
GO:0072503 |
cellular divalent inorganic cation homeostasis(GO:0072503) |
| 0.0 |
0.0 |
GO:0006700 |
C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 |
0.1 |
GO:0046968 |
peptide antigen transport(GO:0046968) |
| 0.0 |
0.3 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.0 |
0.3 |
GO:0007031 |
peroxisome organization(GO:0007031) |
| 0.0 |
0.1 |
GO:0045616 |
regulation of keratinocyte differentiation(GO:0045616) |
| 0.0 |
0.1 |
GO:0006198 |
cAMP catabolic process(GO:0006198) |
| 0.0 |
0.3 |
GO:0008633 |
obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.0 |
0.1 |
GO:0040023 |
establishment of nucleus localization(GO:0040023) |
| 0.0 |
0.7 |
GO:0030833 |
regulation of actin filament polymerization(GO:0030833) |
| 0.0 |
0.1 |
GO:0071636 |
positive regulation of transforming growth factor beta production(GO:0071636) |
| 0.0 |
0.2 |
GO:0071804 |
cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 |
0.2 |
GO:0048709 |
oligodendrocyte differentiation(GO:0048709) |
| 0.0 |
0.2 |
GO:0051568 |
histone H3-K4 methylation(GO:0051568) |
| 0.0 |
0.1 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.2 |
GO:0007507 |
heart development(GO:0007507) |
| 0.0 |
1.7 |
GO:0006412 |
translation(GO:0006412) |
| 0.0 |
0.4 |
GO:0022904 |
respiratory electron transport chain(GO:0022904) |
| 0.0 |
0.0 |
GO:0010628 |
positive regulation of gene expression(GO:0010628) |
| 0.0 |
0.1 |
GO:0035587 |
purinergic receptor signaling pathway(GO:0035587) purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 |
0.6 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
0.1 |
GO:0006612 |
protein targeting to membrane(GO:0006612) |
| 0.0 |
0.0 |
GO:0060972 |
left/right pattern formation(GO:0060972) |