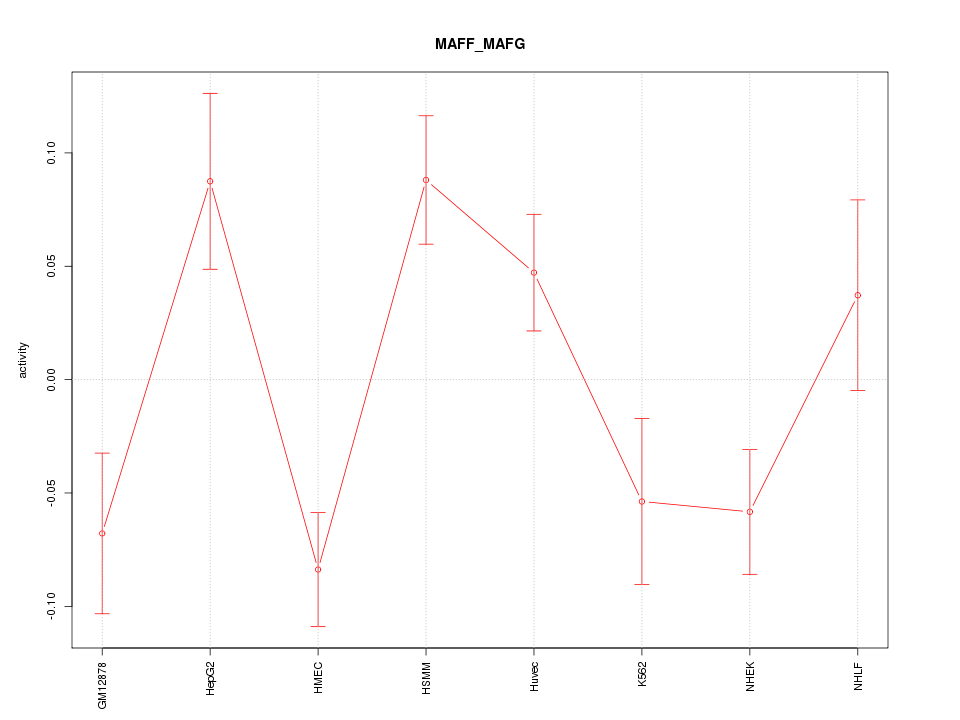
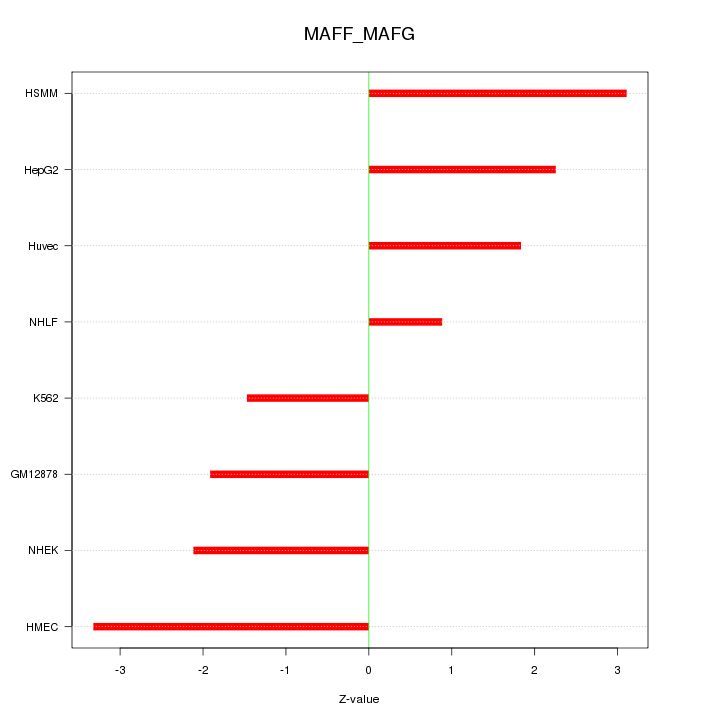
Motif ID: MAFF_MAFG
Z-value: 2.243
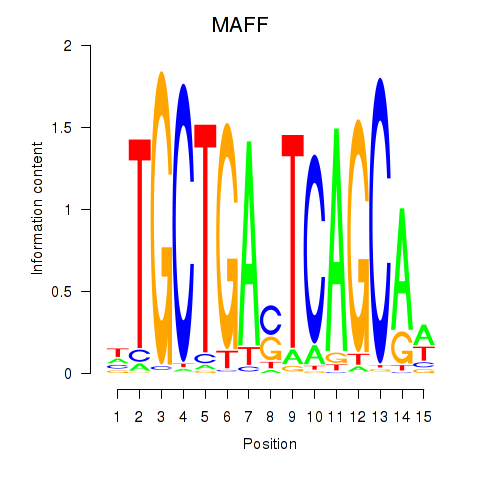
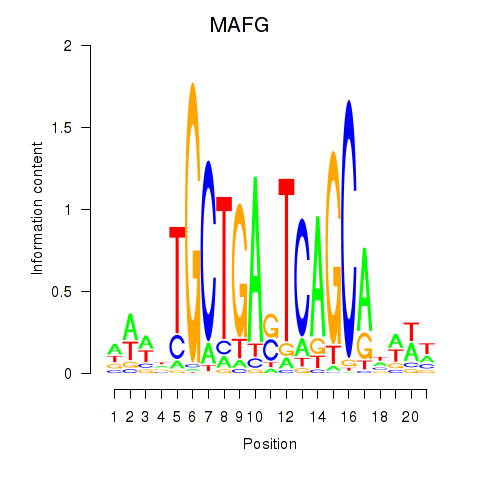
Transcription factors associated with MAFF_MAFG:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| MAFF | ENSG00000185022.7 | MAFF |
| MAFG | ENSG00000197063.6 | MAFG |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0001907 | cytolysis by symbiont of host cells(GO:0001897) killing by symbiont of host cells(GO:0001907) hemolysis by symbiont of host erythrocytes(GO:0019836) disruption by symbiont of host cell(GO:0044004) hemolysis in other organism(GO:0044179) positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) cytolysis in other organism(GO:0051715) cytolysis in other organism involved in symbiotic interaction(GO:0051801) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.6 | 2.5 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.3 | 1.4 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.3 | 0.9 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.3 | 0.9 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.3 | 0.8 | GO:0071883 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.2 | 1.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.2 | 2.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 | 1.3 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.2 | 1.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.2 | 0.9 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.2 | 26.6 | GO:0006415 | translational termination(GO:0006415) |
| 0.2 | 0.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 1.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 1.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 3.4 | GO:0051923 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) sulfation(GO:0051923) |
| 0.1 | 0.5 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.1 | 0.8 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 0.4 | GO:0019448 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) taurine metabolic process(GO:0019530) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 0.7 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.1 | 0.7 | GO:0000050 | urea cycle(GO:0000050) |
| 0.1 | 0.2 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.1 | 0.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.9 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.1 | 0.7 | GO:0006689 | ganglioside catabolic process(GO:0006689) neuromuscular process controlling posture(GO:0050884) |
| 0.1 | 0.2 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.1 | 2.1 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.1 | 0.2 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 0.7 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.9 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.1 | 1.1 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.1 | 0.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.6 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) sensory perception of umami taste(GO:0050917) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.5 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 1.1 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.8 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.3 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0035269 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.1 | GO:0010737 | protein kinase A signaling(GO:0010737) regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.5 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.3 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.5 | GO:0061025 | membrane fusion(GO:0061025) |
| 0.0 | 0.1 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.2 | GO:0046051 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.0 | 0.3 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.4 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.2 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.9 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 3.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:0033866 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 1.7 | GO:0030814 | regulation of cAMP metabolic process(GO:0030814) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.7 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.1 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.0 | 0.4 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 1.1 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.3 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.5 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.0 | 0.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 1.0 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.0 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 1.9 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 3.7 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.3 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.4 | GO:0046464 | triglyceride catabolic process(GO:0019433) neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 1.4 | GO:0048511 | rhythmic process(GO:0048511) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 27.0 | GO:0005844 | polysome(GO:0005844) |
| 0.4 | 1.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 0.6 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 1.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 11.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.6 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 3.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.7 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 2.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.7 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 1.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 2.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.1 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle(GO:0030990) intraciliary transport particle B(GO:0030992) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 26.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.5 | 2.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.4 | 1.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.3 | 0.8 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.3 | 0.8 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.3 | 0.8 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.2 | 3.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 0.9 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 2.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 1.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 2.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 4.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 1.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 0.7 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.2 | 0.5 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.1 | 0.9 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 2.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.4 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.5 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.1 | 0.4 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 0.9 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 1.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.3 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.1 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.3 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.9 | GO:0030547 | receptor inhibitor activity(GO:0030547) receptor antagonist activity(GO:0048019) |
| 0.1 | 2.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.3 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.1 | 0.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.1 | 2.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 3.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.5 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.1 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.5 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.9 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.8 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 0.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.5 | GO:0051637 | obsolete Gram-positive bacterial cell surface binding(GO:0051637) |
| 0.0 | 0.1 | GO:0004608 | phosphatidylethanolamine N-methyltransferase activity(GO:0004608) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 1.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 3.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 1.3 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 1.1 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.4 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.9 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.2 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.0 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.2 | ST_GA13_PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.9 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |