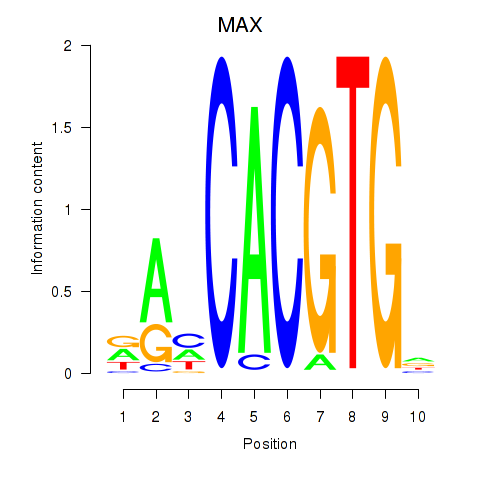
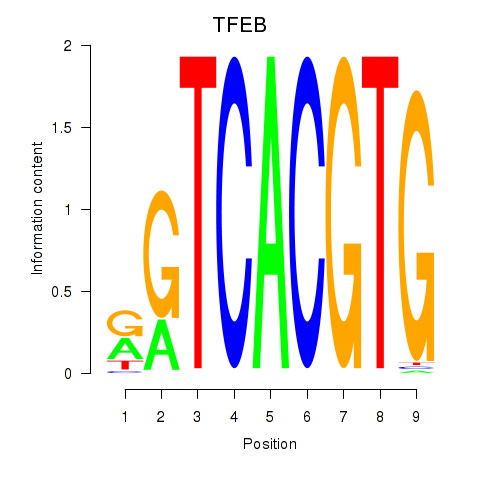
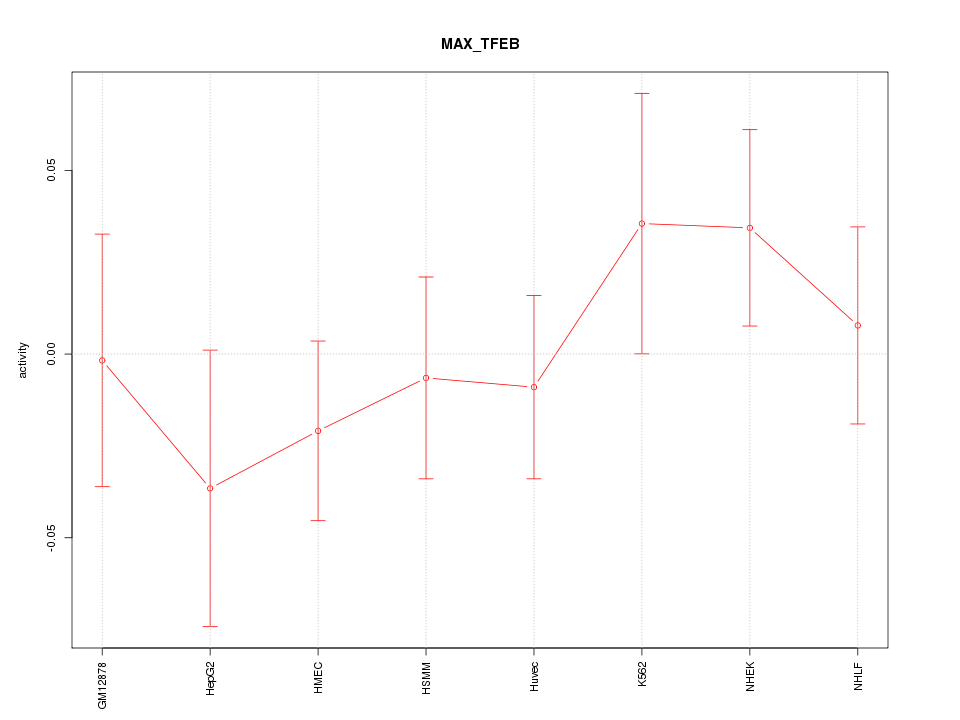
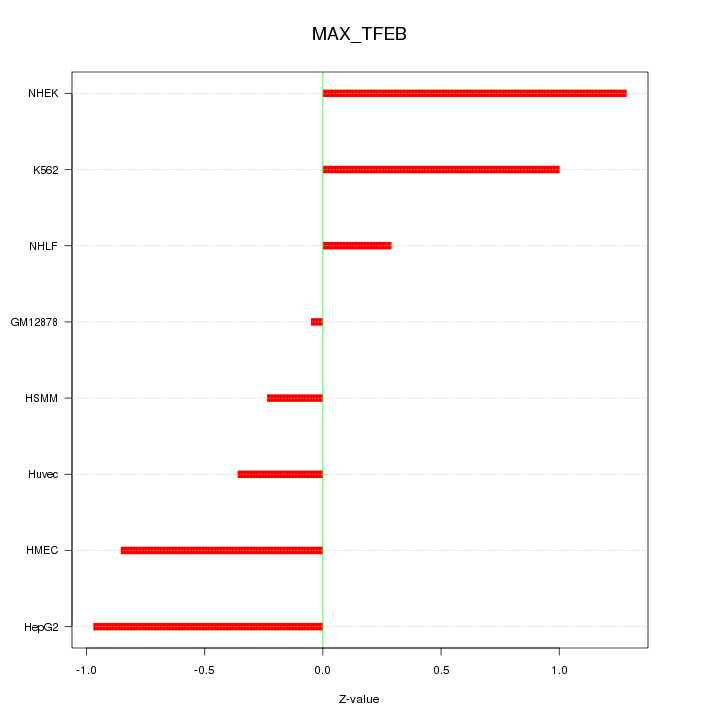
| 0.4 |
2.1 |
GO:0008588 |
release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.2 |
0.7 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.2 |
3.0 |
GO:0048268 |
negative regulation of receptor-mediated endocytosis(GO:0048261) clathrin coat assembly(GO:0048268) |
| 0.2 |
0.5 |
GO:0010586 |
miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.2 |
0.5 |
GO:0046449 |
allantoin metabolic process(GO:0000255) mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.2 |
0.5 |
GO:0031118 |
rRNA pseudouridine synthesis(GO:0031118) |
| 0.2 |
0.8 |
GO:0032933 |
SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 |
1.0 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.1 |
2.6 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 |
0.4 |
GO:0021570 |
rhombomere 4 development(GO:0021570) facial nucleus development(GO:0021754) |
| 0.1 |
1.0 |
GO:0071243 |
uroporphyrinogen III metabolic process(GO:0046502) response to fungicide(GO:0060992) cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 |
0.4 |
GO:0009202 |
purine deoxyribonucleotide biosynthetic process(GO:0009153) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.1 |
0.4 |
GO:0046604 |
regulation of mitotic centrosome separation(GO:0046602) positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 |
0.5 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.1 |
0.5 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.1 |
1.2 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 |
0.3 |
GO:0032196 |
transposition, DNA-mediated(GO:0006313) transposition(GO:0032196) |
| 0.1 |
0.4 |
GO:0010944 |
negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.1 |
0.4 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.1 |
0.3 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 |
0.4 |
GO:0009698 |
phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.1 |
0.8 |
GO:0045008 |
depyrimidination(GO:0045008) |
| 0.1 |
0.1 |
GO:0021898 |
commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.1 |
3.8 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 |
0.3 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 |
0.2 |
GO:0034625 |
fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.1 |
0.6 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 |
0.3 |
GO:0001575 |
globoside metabolic process(GO:0001575) |
| 0.1 |
0.8 |
GO:0000076 |
DNA replication checkpoint(GO:0000076) |
| 0.1 |
0.3 |
GO:0019348 |
polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.1 |
0.2 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.1 |
2.0 |
GO:0044349 |
nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) |
| 0.1 |
0.2 |
GO:0015670 |
carbon dioxide transport(GO:0015670) |
| 0.1 |
0.6 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.1 |
1.1 |
GO:0006596 |
polyamine biosynthetic process(GO:0006596) |
| 0.1 |
0.9 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.1 |
0.5 |
GO:0006474 |
N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 |
0.4 |
GO:0007021 |
tubulin complex assembly(GO:0007021) acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.1 |
0.2 |
GO:0043457 |
regulation of cellular respiration(GO:0043457) |
| 0.1 |
0.1 |
GO:0045618 |
positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 |
2.6 |
GO:0048146 |
positive regulation of fibroblast proliferation(GO:0048146) |
| 0.1 |
0.4 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 |
0.2 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 |
0.2 |
GO:0090467 |
arginine transport(GO:0015809) L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.1 |
0.2 |
GO:0010520 |
regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) regulation of meiosis I(GO:0060631) |
| 0.0 |
0.4 |
GO:0006188 |
IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.0 |
0.5 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.0 |
0.3 |
GO:0060528 |
secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 |
0.3 |
GO:1901984 |
negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 |
0.2 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.2 |
GO:0060649 |
nipple development(GO:0060618) mammary gland bud elongation(GO:0060649) nipple morphogenesis(GO:0060658) nipple sheath formation(GO:0060659) |
| 0.0 |
0.1 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 |
1.6 |
GO:0016925 |
protein sumoylation(GO:0016925) |
| 0.0 |
0.6 |
GO:0097031 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.1 |
GO:0002541 |
activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 |
0.1 |
GO:0045656 |
negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 |
0.6 |
GO:0006044 |
N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 |
0.7 |
GO:0048169 |
regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 |
0.5 |
GO:0055015 |
ventricular cardiac muscle cell development(GO:0055015) |
| 0.0 |
0.2 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 |
0.3 |
GO:0034755 |
iron ion transmembrane transport(GO:0034755) |
| 0.0 |
0.3 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.0 |
0.6 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.5 |
GO:0032703 |
negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 |
1.0 |
GO:0034199 |
activation of protein kinase A activity(GO:0034199) |
| 0.0 |
0.1 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
0.8 |
GO:0035196 |
production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 |
0.4 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.7 |
GO:0000380 |
alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 |
0.2 |
GO:0060259 |
regulation of feeding behavior(GO:0060259) |
| 0.0 |
0.1 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.0 |
1.2 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.1 |
GO:0060896 |
neural plate anterior/posterior regionalization(GO:0021999) neural plate pattern specification(GO:0060896) neural plate regionalization(GO:0060897) |
| 0.0 |
0.2 |
GO:1903301 |
regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 |
0.2 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 |
0.1 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 |
0.1 |
GO:0070384 |
Harderian gland development(GO:0070384) |
| 0.0 |
0.2 |
GO:0015904 |
tetracycline transport(GO:0015904) |
| 0.0 |
0.4 |
GO:0046685 |
response to arsenic-containing substance(GO:0046685) |
| 0.0 |
0.1 |
GO:0072387 |
flavin-containing compound biosynthetic process(GO:0042727) flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.0 |
0.3 |
GO:0051290 |
protein heterotetramerization(GO:0051290) |
| 0.0 |
0.7 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.3 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.0 |
0.3 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |
| 0.0 |
0.2 |
GO:0022614 |
membrane to membrane docking(GO:0022614) |
| 0.0 |
0.3 |
GO:0032647 |
interferon-alpha production(GO:0032607) regulation of interferon-alpha production(GO:0032647) positive regulation of interferon-alpha production(GO:0032727) |
| 0.0 |
0.1 |
GO:0070734 |
histone H3-K27 methylation(GO:0070734) |
| 0.0 |
0.3 |
GO:0032481 |
positive regulation of type I interferon production(GO:0032481) positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.2 |
GO:0003310 |
pancreatic A cell differentiation(GO:0003310) |
| 0.0 |
0.3 |
GO:0098927 |
early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 |
0.1 |
GO:0033160 |
positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 |
0.1 |
GO:0035195 |
gene silencing by miRNA(GO:0035195) |
| 0.0 |
0.2 |
GO:0007080 |
mitotic metaphase plate congression(GO:0007080) |
| 0.0 |
0.2 |
GO:0006552 |
leucine catabolic process(GO:0006552) |
| 0.0 |
0.1 |
GO:0034723 |
DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
0.1 |
GO:2000124 |
cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 |
0.2 |
GO:0045898 |
regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 |
0.6 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 |
0.1 |
GO:0046108 |
uridine metabolic process(GO:0046108) |
| 0.0 |
0.4 |
GO:0035036 |
binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 |
1.0 |
GO:0000289 |
nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 |
2.1 |
GO:0006364 |
rRNA processing(GO:0006364) |
| 0.0 |
0.2 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.1 |
GO:1901798 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 |
0.2 |
GO:0006069 |
ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 |
0.4 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.1 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 |
0.3 |
GO:0030500 |
regulation of bone mineralization(GO:0030500) |
| 0.0 |
0.3 |
GO:0009303 |
rRNA transcription(GO:0009303) |
| 0.0 |
0.4 |
GO:0048704 |
embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 |
0.0 |
GO:0031580 |
membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 |
0.2 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 |
0.5 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.0 |
0.1 |
GO:0006047 |
UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 |
1.3 |
GO:0006892 |
post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 |
0.2 |
GO:0035246 |
peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 |
1.6 |
GO:0048193 |
Golgi vesicle transport(GO:0048193) |
| 0.0 |
0.2 |
GO:0006546 |
glycine catabolic process(GO:0006546) |
| 0.0 |
0.1 |
GO:0019056 |
modulation by virus of host transcription(GO:0019056) regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 |
0.3 |
GO:0006400 |
tRNA modification(GO:0006400) |
| 0.0 |
0.4 |
GO:0006099 |
tricarboxylic acid cycle(GO:0006099) |
| 0.0 |
0.1 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.0 |
0.1 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.0 |
0.1 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 |
0.1 |
GO:0043045 |
DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 |
0.5 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 |
0.1 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 |
0.3 |
GO:0006490 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 |
0.2 |
GO:0051568 |
histone H3-K4 methylation(GO:0051568) |
| 0.0 |
0.6 |
GO:0042254 |
ribosome biogenesis(GO:0042254) |
| 0.0 |
0.2 |
GO:0060351 |
cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 |
0.4 |
GO:0048255 |
RNA stabilization(GO:0043489) mRNA stabilization(GO:0048255) |
| 0.0 |
0.1 |
GO:0015853 |
adenine transport(GO:0015853) |
| 0.0 |
0.1 |
GO:0033572 |
ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 |
0.3 |
GO:0008333 |
endosome to lysosome transport(GO:0008333) |
| 0.0 |
0.4 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.0 |
0.1 |
GO:1901343 |
negative regulation of angiogenesis(GO:0016525) negative regulation of vasculature development(GO:1901343) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.0 |
0.1 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.3 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.1 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.0 |
0.2 |
GO:0045670 |
regulation of osteoclast differentiation(GO:0045670) |
| 0.0 |
0.1 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.2 |
GO:0060216 |
definitive hemopoiesis(GO:0060216) |
| 0.0 |
0.1 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
0.1 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 |
0.1 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.0 |
0.2 |
GO:0006098 |
pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 |
0.1 |
GO:0000186 |
activation of MAPKK activity(GO:0000186) |
| 0.0 |
0.1 |
GO:0002724 |
T cell cytokine production(GO:0002369) regulation of T cell cytokine production(GO:0002724) positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 |
0.1 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.0 |
0.2 |
GO:0031397 |
negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 |
0.0 |
GO:0006382 |
adenosine to inosine editing(GO:0006382) |
| 0.0 |
0.1 |
GO:0000731 |
DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 |
0.0 |
GO:0021615 |
glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 |
0.2 |
GO:0046902 |
regulation of mitochondrial membrane permeability(GO:0046902) regulation of membrane permeability(GO:0090559) |
| 0.0 |
0.0 |
GO:0002467 |
germinal center formation(GO:0002467) |
| 0.0 |
0.4 |
GO:0006612 |
protein targeting to membrane(GO:0006612) |
| 0.0 |
0.1 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 |
0.0 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 |
0.0 |
GO:0010273 |
regulation of oxidative phosphorylation(GO:0002082) detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 |
0.7 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.1 |
GO:0042977 |
activation of JAK2 kinase activity(GO:0042977) |
| 0.0 |
0.1 |
GO:0048070 |
regulation of developmental pigmentation(GO:0048070) |
| 0.0 |
0.1 |
GO:0006903 |
vesicle targeting(GO:0006903) |