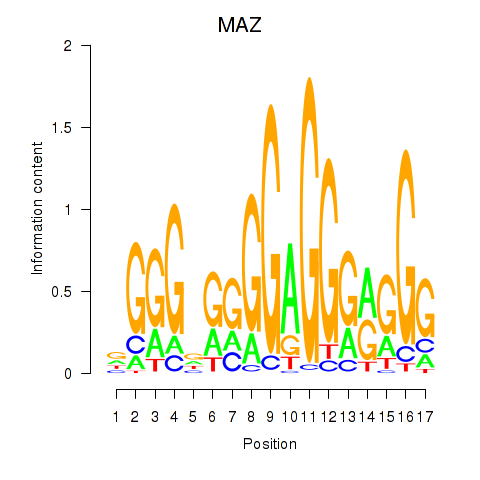
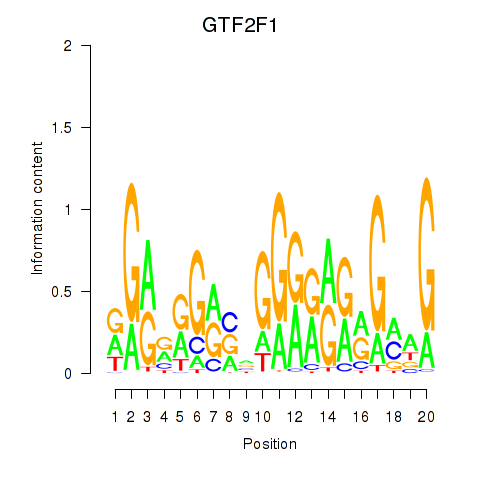
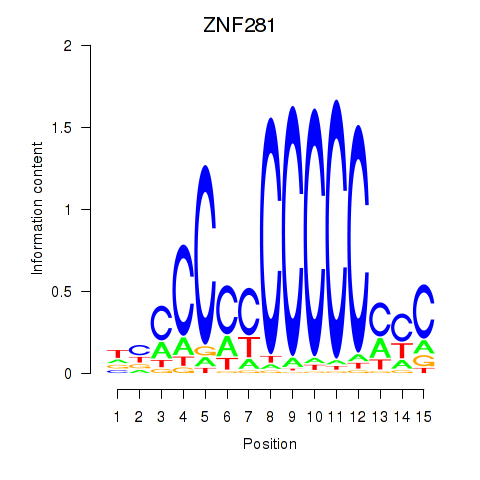
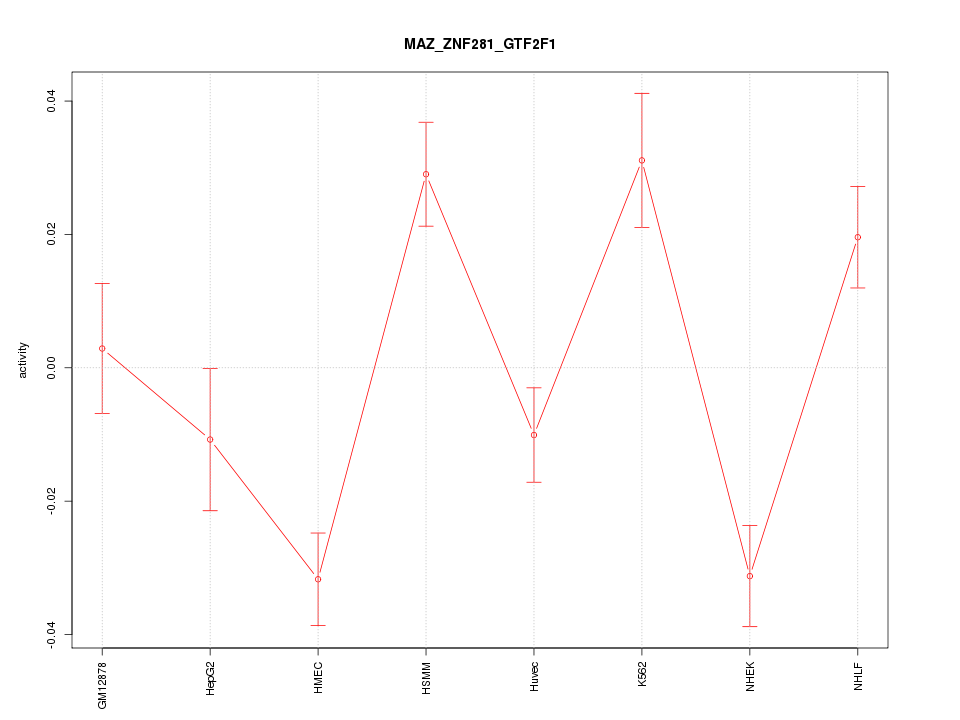
| 2.1 |
8.3 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 1.8 |
7.1 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 1.0 |
8.1 |
GO:0031444 |
slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.9 |
2.7 |
GO:0061030 |
epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.8 |
2.5 |
GO:0035408 |
histone H3-T6 phosphorylation(GO:0035408) |
| 0.7 |
2.1 |
GO:0043371 |
negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.7 |
0.7 |
GO:0043113 |
receptor clustering(GO:0043113) |
| 0.7 |
2.0 |
GO:0010586 |
miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.7 |
2.0 |
GO:0043418 |
cysteine biosynthetic process from serine(GO:0006535) homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) transsulfuration(GO:0019346) homocysteine catabolic process(GO:0043418) |
| 0.7 |
3.3 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.5 |
1.1 |
GO:0071786 |
endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.5 |
10.6 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.5 |
2.0 |
GO:0051573 |
negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.5 |
0.5 |
GO:0048048 |
embryonic eye morphogenesis(GO:0048048) |
| 0.5 |
1.4 |
GO:0021919 |
BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.5 |
1.4 |
GO:0060599 |
lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.5 |
0.5 |
GO:0060666 |
dichotomous subdivision of an epithelial terminal unit(GO:0060600) dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.4 |
2.2 |
GO:0003093 |
regulation of glomerular filtration(GO:0003093) glomerular filtration(GO:0003094) renal filtration(GO:0097205) regulation of renal system process(GO:0098801) |
| 0.4 |
1.3 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) |
| 0.4 |
1.8 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.4 |
11.0 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.4 |
1.7 |
GO:0000189 |
MAPK import into nucleus(GO:0000189) |
| 0.4 |
2.1 |
GO:0032696 |
negative regulation of interleukin-13 production(GO:0032696) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.4 |
2.9 |
GO:0030007 |
cellular potassium ion homeostasis(GO:0030007) negative regulation of cell volume(GO:0045794) |
| 0.4 |
1.2 |
GO:0006679 |
glucosylceramide biosynthetic process(GO:0006679) |
| 0.4 |
1.2 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.4 |
1.1 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.4 |
1.5 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.4 |
0.8 |
GO:0051205 |
protein insertion into membrane(GO:0051205) |
| 0.4 |
2.3 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.4 |
8.7 |
GO:0021846 |
cell proliferation in forebrain(GO:0021846) |
| 0.4 |
2.2 |
GO:0030240 |
skeletal muscle thin filament assembly(GO:0030240) |
| 0.4 |
1.1 |
GO:2000105 |
positive regulation of DNA-dependent DNA replication initiation(GO:0032298) positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.4 |
1.1 |
GO:1904036 |
negative regulation of epithelial cell apoptotic process(GO:1904036) negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.3 |
3.1 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.3 |
1.0 |
GO:0090047 |
obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.3 |
1.4 |
GO:0071313 |
cellular response to caffeine(GO:0071313) |
| 0.3 |
1.0 |
GO:0071529 |
cementum mineralization(GO:0071529) |
| 0.3 |
1.0 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.3 |
0.9 |
GO:0001544 |
initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.3 |
1.8 |
GO:0009082 |
branched-chain amino acid biosynthetic process(GO:0009082) |
| 0.3 |
0.9 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.3 |
1.5 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.3 |
1.2 |
GO:0045199 |
maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.3 |
0.6 |
GO:0000414 |
regulation of histone H3-K36 methylation(GO:0000414) |
| 0.3 |
1.2 |
GO:0006089 |
lactate metabolic process(GO:0006089) |
| 0.3 |
1.8 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.3 |
2.9 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.3 |
0.9 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.3 |
4.3 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.3 |
0.3 |
GO:0045992 |
negative regulation of embryonic development(GO:0045992) |
| 0.3 |
1.7 |
GO:0032916 |
transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.3 |
1.7 |
GO:0055009 |
atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.3 |
3.9 |
GO:0006527 |
arginine catabolic process(GO:0006527) |
| 0.3 |
0.8 |
GO:0002158 |
osteoclast proliferation(GO:0002158) |
| 0.3 |
0.8 |
GO:0000114 |
obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.3 |
1.6 |
GO:0030186 |
melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.3 |
0.5 |
GO:0048263 |
determination of dorsal identity(GO:0048263) |
| 0.3 |
0.3 |
GO:0060279 |
regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.3 |
0.5 |
GO:0060462 |
lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.3 |
0.8 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.3 |
0.5 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.3 |
8.8 |
GO:0030325 |
adrenal gland development(GO:0030325) |
| 0.3 |
0.5 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.3 |
2.9 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 |
1.1 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.3 |
1.0 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.3 |
0.8 |
GO:0046015 |
carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.3 |
1.8 |
GO:0042483 |
negative regulation of odontogenesis(GO:0042483) |
| 0.3 |
1.5 |
GO:0040037 |
negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.3 |
1.0 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.3 |
1.0 |
GO:0002318 |
myeloid progenitor cell differentiation(GO:0002318) |
| 0.2 |
0.2 |
GO:0035413 |
positive regulation of catenin import into nucleus(GO:0035413) |
| 0.2 |
1.2 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.2 |
0.2 |
GO:1902806 |
regulation of cell cycle G1/S phase transition(GO:1902806) regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.2 |
3.1 |
GO:0031498 |
nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.2 |
0.7 |
GO:0071280 |
epithelial fluid transport(GO:0042045) cellular response to copper ion(GO:0071280) |
| 0.2 |
0.7 |
GO:0060318 |
definitive erythrocyte differentiation(GO:0060318) |
| 0.2 |
5.0 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.2 |
0.7 |
GO:0048369 |
lateral mesoderm development(GO:0048368) lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.2 |
0.7 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.2 |
0.7 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 |
1.4 |
GO:0014819 |
regulation of skeletal muscle contraction(GO:0014819) |
| 0.2 |
2.5 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 |
1.5 |
GO:0051895 |
negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.2 |
0.4 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.2 |
1.1 |
GO:0006857 |
oligopeptide transport(GO:0006857) |
| 0.2 |
0.7 |
GO:0034036 |
purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 |
0.7 |
GO:0007612 |
learning(GO:0007612) |
| 0.2 |
6.7 |
GO:0060021 |
palate development(GO:0060021) |
| 0.2 |
0.2 |
GO:2000648 |
positive regulation of stem cell proliferation(GO:2000648) |
| 0.2 |
1.1 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.2 |
0.6 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) |
| 0.2 |
0.6 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.2 |
19.9 |
GO:0009408 |
response to heat(GO:0009408) |
| 0.2 |
1.2 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.2 |
0.8 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.2 |
0.4 |
GO:0051971 |
positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.2 |
0.8 |
GO:0032354 |
response to follicle-stimulating hormone(GO:0032354) |
| 0.2 |
1.0 |
GO:0060014 |
granulosa cell differentiation(GO:0060014) |
| 0.2 |
1.2 |
GO:0010759 |
positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.2 |
4.8 |
GO:0045103 |
intermediate filament-based process(GO:0045103) |
| 0.2 |
0.8 |
GO:0010900 |
regulation of phosphatidylcholine catabolic process(GO:0010899) negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.2 |
0.9 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 |
0.6 |
GO:0021978 |
telencephalon regionalization(GO:0021978) |
| 0.2 |
0.7 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 |
0.6 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.2 |
1.6 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.2 |
0.9 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.2 |
0.2 |
GO:0035265 |
organ growth(GO:0035265) |
| 0.2 |
1.8 |
GO:0021877 |
forebrain neuron fate commitment(GO:0021877) |
| 0.2 |
0.2 |
GO:0072202 |
cell differentiation involved in metanephros development(GO:0072202) |
| 0.2 |
0.5 |
GO:0034770 |
histone H4-K20 methylation(GO:0034770) |
| 0.2 |
1.0 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 |
0.5 |
GO:2000121 |
regulation of removal of superoxide radicals(GO:2000121) |
| 0.2 |
0.3 |
GO:0042984 |
amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 |
3.4 |
GO:0008347 |
glial cell migration(GO:0008347) |
| 0.2 |
1.2 |
GO:0042117 |
monocyte activation(GO:0042117) |
| 0.2 |
0.9 |
GO:0006942 |
regulation of striated muscle contraction(GO:0006942) |
| 0.2 |
0.3 |
GO:0045835 |
negative regulation of meiotic nuclear division(GO:0045835) |
| 0.2 |
0.7 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.2 |
0.3 |
GO:0060394 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 |
0.3 |
GO:2000644 |
low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.2 |
0.5 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.2 |
0.2 |
GO:0009756 |
carbohydrate mediated signaling(GO:0009756) |
| 0.2 |
0.6 |
GO:0045900 |
negative regulation of translational elongation(GO:0045900) |
| 0.2 |
0.5 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.2 |
0.3 |
GO:0006154 |
adenosine catabolic process(GO:0006154) |
| 0.2 |
1.4 |
GO:1900087 |
traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.2 |
0.8 |
GO:0051024 |
positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.2 |
0.9 |
GO:0048537 |
mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.2 |
0.3 |
GO:0033632 |
regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 |
1.3 |
GO:0002092 |
positive regulation of receptor internalization(GO:0002092) |
| 0.2 |
0.5 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.2 |
1.1 |
GO:0060770 |
negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.2 |
0.2 |
GO:0003032 |
detection of oxygen(GO:0003032) |
| 0.2 |
0.5 |
GO:0007206 |
phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 |
1.2 |
GO:0033261 |
obsolete regulation of S phase(GO:0033261) |
| 0.2 |
0.5 |
GO:0050758 |
thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.2 |
0.6 |
GO:2000189 |
regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.2 |
0.6 |
GO:0006768 |
biotin metabolic process(GO:0006768) |
| 0.1 |
0.6 |
GO:0060710 |
chorio-allantoic fusion(GO:0060710) |
| 0.1 |
0.6 |
GO:0031635 |
adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) opioid receptor signaling pathway(GO:0038003) |
| 0.1 |
0.1 |
GO:0009648 |
photoperiodism(GO:0009648) |
| 0.1 |
1.0 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.1 |
0.9 |
GO:0035058 |
nonmotile primary cilium assembly(GO:0035058) |
| 0.1 |
0.4 |
GO:0046292 |
formaldehyde metabolic process(GO:0046292) |
| 0.1 |
1.0 |
GO:0042693 |
muscle cell fate commitment(GO:0042693) |
| 0.1 |
0.7 |
GO:0030500 |
regulation of bone mineralization(GO:0030500) |
| 0.1 |
0.6 |
GO:0010829 |
negative regulation of glucose transport(GO:0010829) |
| 0.1 |
0.1 |
GO:0036037 |
CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 |
0.3 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.1 |
1.1 |
GO:0051383 |
kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.1 |
0.1 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 |
0.3 |
GO:0008628 |
hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.1 |
0.4 |
GO:0015854 |
guanine transport(GO:0015854) hypoxanthine transport(GO:0035344) thymine transport(GO:0035364) |
| 0.1 |
1.4 |
GO:0050847 |
progesterone receptor signaling pathway(GO:0050847) |
| 0.1 |
0.4 |
GO:0034616 |
response to laminar fluid shear stress(GO:0034616) |
| 0.1 |
0.1 |
GO:0001553 |
luteinization(GO:0001553) progesterone secretion(GO:0042701) |
| 0.1 |
0.8 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.1 |
0.7 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.1 |
0.7 |
GO:0001837 |
epithelial to mesenchymal transition(GO:0001837) |
| 0.1 |
0.4 |
GO:0051712 |
positive regulation of killing of cells of other organism(GO:0051712) |
| 0.1 |
0.4 |
GO:0070384 |
Harderian gland development(GO:0070384) |
| 0.1 |
0.7 |
GO:0031274 |
pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 |
0.1 |
GO:1903509 |
liposaccharide metabolic process(GO:1903509) |
| 0.1 |
0.6 |
GO:0001880 |
Mullerian duct regression(GO:0001880) |
| 0.1 |
0.8 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 |
0.5 |
GO:0021517 |
ventral spinal cord development(GO:0021517) spinal cord motor neuron differentiation(GO:0021522) |
| 0.1 |
0.5 |
GO:0009296 |
obsolete flagellum assembly(GO:0009296) |
| 0.1 |
0.4 |
GO:0002903 |
negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 |
1.0 |
GO:0046831 |
regulation of RNA export from nucleus(GO:0046831) |
| 0.1 |
1.4 |
GO:0009148 |
pyrimidine nucleoside triphosphate biosynthetic process(GO:0009148) |
| 0.1 |
0.4 |
GO:0015959 |
diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.1 |
4.1 |
GO:0043462 |
regulation of ATPase activity(GO:0043462) |
| 0.1 |
1.3 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 |
0.9 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.1 |
0.7 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.1 |
1.7 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 |
2.5 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.1 |
0.7 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 |
1.3 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.1 |
0.7 |
GO:0001514 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 |
0.1 |
GO:0010828 |
positive regulation of glucose transport(GO:0010828) |
| 0.1 |
1.3 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 |
0.4 |
GO:0033211 |
adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 |
0.1 |
GO:0046619 |
optic placode formation(GO:0001743) optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 |
0.6 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 |
0.1 |
GO:0048861 |
leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 |
0.4 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.1 |
0.8 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.1 |
0.3 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.1 |
0.2 |
GO:0006307 |
DNA dealkylation involved in DNA repair(GO:0006307) DNA dealkylation(GO:0035510) |
| 0.1 |
0.2 |
GO:0046732 |
induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by virus of host immune response(GO:0075528) |
| 0.1 |
0.3 |
GO:0018916 |
nitrobenzene metabolic process(GO:0018916) |
| 0.1 |
0.3 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 |
0.6 |
GO:0051154 |
negative regulation of myotube differentiation(GO:0010832) negative regulation of striated muscle cell differentiation(GO:0051154) |
| 0.1 |
0.2 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.3 |
GO:0019371 |
cyclooxygenase pathway(GO:0019371) |
| 0.1 |
0.9 |
GO:1901985 |
positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.1 |
0.8 |
GO:0071243 |
cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 |
0.8 |
GO:0048024 |
regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.1 |
0.8 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.1 |
0.5 |
GO:0042474 |
middle ear morphogenesis(GO:0042474) |
| 0.1 |
0.1 |
GO:0006553 |
lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.1 |
0.5 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 |
0.6 |
GO:0060219 |
camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 |
0.3 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 |
0.9 |
GO:0015705 |
iodide transport(GO:0015705) |
| 0.1 |
0.9 |
GO:0048641 |
regulation of skeletal muscle tissue development(GO:0048641) |
| 0.1 |
0.6 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.1 |
0.2 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.1 |
0.8 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.1 |
0.1 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.1 |
0.2 |
GO:0033137 |
negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.1 |
0.1 |
GO:0002678 |
positive regulation of chronic inflammatory response(GO:0002678) |
| 0.1 |
0.7 |
GO:0090090 |
negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 |
0.2 |
GO:0010520 |
regulation of reciprocal meiotic recombination(GO:0010520) regulation of meiosis I(GO:0060631) |
| 0.1 |
1.0 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.1 |
0.3 |
GO:0098781 |
rRNA transcription(GO:0009303) ncRNA transcription(GO:0098781) |
| 0.1 |
0.6 |
GO:0010523 |
negative regulation of calcium ion transport into cytosol(GO:0010523) |
| 0.1 |
3.1 |
GO:0030866 |
cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 |
0.2 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.1 |
0.4 |
GO:0046874 |
quinolinate metabolic process(GO:0046874) |
| 0.1 |
0.1 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.1 |
0.1 |
GO:0034976 |
response to endoplasmic reticulum stress(GO:0034976) |
| 0.1 |
1.8 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 |
1.7 |
GO:0075713 |
establishment of viral latency(GO:0019043) establishment of integrated proviral latency(GO:0075713) |
| 0.1 |
2.0 |
GO:0065004 |
protein-DNA complex assembly(GO:0065004) |
| 0.1 |
0.3 |
GO:0006782 |
protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.1 |
0.3 |
GO:0000338 |
protein deneddylation(GO:0000338) |
| 0.1 |
0.3 |
GO:1990170 |
detoxification of cadmium ion(GO:0071585) stress response to cadmium ion(GO:1990170) |
| 0.1 |
2.6 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.1 |
0.5 |
GO:0021520 |
spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 |
0.1 |
GO:0060029 |
convergent extension involved in organogenesis(GO:0060029) |
| 0.1 |
0.4 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 |
0.2 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 |
0.1 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.1 |
0.3 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 |
0.2 |
GO:0060033 |
anatomical structure regression(GO:0060033) |
| 0.1 |
0.3 |
GO:0051901 |
positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.1 |
0.1 |
GO:2000078 |
regulation of type B pancreatic cell development(GO:2000074) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.1 |
0.1 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.1 |
0.5 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 |
0.5 |
GO:0008354 |
germ cell migration(GO:0008354) |
| 0.1 |
0.4 |
GO:0043128 |
regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 |
0.1 |
GO:0043049 |
otic placode formation(GO:0043049) |
| 0.1 |
0.7 |
GO:0046839 |
phospholipid dephosphorylation(GO:0046839) |
| 0.1 |
0.1 |
GO:0060259 |
regulation of feeding behavior(GO:0060259) |
| 0.1 |
0.2 |
GO:0072136 |
kidney mesenchymal cell proliferation(GO:0072135) metanephric mesenchymal cell proliferation involved in metanephros development(GO:0072136) |
| 0.1 |
0.4 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 |
0.5 |
GO:0043316 |
natural killer cell activation involved in immune response(GO:0002323) cytotoxic T cell degranulation(GO:0043316) natural killer cell degranulation(GO:0043320) |
| 0.1 |
0.2 |
GO:0001569 |
patterning of blood vessels(GO:0001569) |
| 0.1 |
0.3 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.1 |
0.3 |
GO:0046600 |
negative regulation of centriole replication(GO:0046600) negative regulation of organelle assembly(GO:1902116) |
| 0.1 |
2.0 |
GO:0035137 |
hindlimb morphogenesis(GO:0035137) |
| 0.1 |
0.2 |
GO:0090274 |
positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 |
0.3 |
GO:0009153 |
purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.1 |
0.1 |
GO:2000178 |
negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 |
0.3 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.1 |
6.8 |
GO:0008016 |
regulation of heart contraction(GO:0008016) |
| 0.1 |
0.3 |
GO:0006221 |
pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.1 |
0.2 |
GO:0048702 |
embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 |
0.1 |
GO:0098762 |
meiotic cell cycle phase(GO:0098762) meiosis I cell cycle phase(GO:0098764) |
| 0.1 |
0.3 |
GO:0006216 |
cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 |
0.3 |
GO:0020027 |
hemoglobin metabolic process(GO:0020027) |
| 0.1 |
1.8 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.1 |
0.5 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 |
0.3 |
GO:1901419 |
regulation of response to alcohol(GO:1901419) |
| 0.1 |
0.4 |
GO:0031122 |
cytoplasmic microtubule organization(GO:0031122) |
| 0.1 |
0.7 |
GO:0016998 |
cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.1 |
0.1 |
GO:0043931 |
ossification involved in bone maturation(GO:0043931) |
| 0.1 |
0.1 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 |
0.3 |
GO:0045682 |
regulation of epidermis development(GO:0045682) |
| 0.1 |
0.4 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.1 |
0.8 |
GO:0032411 |
positive regulation of transporter activity(GO:0032411) |
| 0.1 |
0.3 |
GO:0001941 |
postsynaptic membrane organization(GO:0001941) |
| 0.1 |
0.1 |
GO:0002385 |
innate immune response in mucosa(GO:0002227) mucosal immune response(GO:0002385) |
| 0.1 |
0.9 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.1 |
3.4 |
GO:0006937 |
regulation of muscle contraction(GO:0006937) |
| 0.1 |
0.1 |
GO:0000183 |
chromatin silencing at rDNA(GO:0000183) |
| 0.1 |
0.4 |
GO:0007289 |
spermatid nucleus differentiation(GO:0007289) |
| 0.1 |
0.2 |
GO:0007285 |
primary spermatocyte growth(GO:0007285) |
| 0.1 |
0.1 |
GO:1903426 |
regulation of nitric oxide biosynthetic process(GO:0045428) regulation of reactive oxygen species biosynthetic process(GO:1903426) |
| 0.1 |
0.9 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.1 |
1.1 |
GO:0060760 |
positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.1 |
0.1 |
GO:0050731 |
positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.1 |
0.1 |
GO:0033135 |
regulation of peptidyl-serine phosphorylation(GO:0033135) |
| 0.1 |
0.9 |
GO:0031000 |
response to caffeine(GO:0031000) |
| 0.1 |
0.2 |
GO:0060574 |
columnar/cuboidal epithelial cell maturation(GO:0002069) transformation of host cell by virus(GO:0019087) intestinal epithelial cell maturation(GO:0060574) intestinal epithelial cell development(GO:0060576) |
| 0.1 |
0.2 |
GO:0016926 |
protein desumoylation(GO:0016926) |
| 0.1 |
0.5 |
GO:0009128 |
AMP catabolic process(GO:0006196) purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.1 |
0.2 |
GO:0014015 |
positive regulation of gliogenesis(GO:0014015) |
| 0.1 |
0.9 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 |
4.3 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 |
0.1 |
GO:0060412 |
ventricular septum morphogenesis(GO:0060412) |
| 0.1 |
0.2 |
GO:0071104 |
negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.1 |
1.6 |
GO:0048255 |
RNA stabilization(GO:0043489) mRNA stabilization(GO:0048255) |
| 0.1 |
1.9 |
GO:0030851 |
granulocyte differentiation(GO:0030851) |
| 0.1 |
0.1 |
GO:0050968 |
detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 |
0.1 |
GO:0014068 |
positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.1 |
0.3 |
GO:0090224 |
regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.1 |
0.6 |
GO:0021511 |
spinal cord patterning(GO:0021511) |
| 0.1 |
0.1 |
GO:0043501 |
skeletal muscle adaptation(GO:0043501) |
| 0.1 |
0.9 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 |
0.4 |
GO:0001782 |
B cell homeostasis(GO:0001782) |
| 0.1 |
0.1 |
GO:0072207 |
metanephric tubule development(GO:0072170) metanephric epithelium development(GO:0072207) metanephric nephron tubule development(GO:0072234) metanephric nephron epithelium development(GO:0072243) |
| 0.1 |
0.1 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.1 |
0.2 |
GO:0070781 |
response to biotin(GO:0070781) |
| 0.1 |
0.9 |
GO:0007498 |
mesoderm development(GO:0007498) |
| 0.1 |
0.5 |
GO:0051001 |
negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 |
0.1 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 |
1.0 |
GO:0007569 |
cell aging(GO:0007569) |
| 0.1 |
0.2 |
GO:0034755 |
iron ion transmembrane transport(GO:0034755) |
| 0.1 |
0.1 |
GO:0060742 |
epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.1 |
2.1 |
GO:0007052 |
mitotic spindle organization(GO:0007052) |
| 0.1 |
0.2 |
GO:0008218 |
bioluminescence(GO:0008218) |
| 0.1 |
0.4 |
GO:0002478 |
antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 0.1 |
1.3 |
GO:0006596 |
polyamine biosynthetic process(GO:0006596) |
| 0.1 |
0.2 |
GO:0035470 |
positive regulation of vascular wound healing(GO:0035470) |
| 0.1 |
0.1 |
GO:0030948 |
negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 |
0.1 |
GO:0031558 |
obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.1 |
0.1 |
GO:0090206 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 |
0.3 |
GO:0071168 |
protein localization to chromatin(GO:0071168) |
| 0.1 |
0.8 |
GO:0009713 |
catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.1 |
0.1 |
GO:0032898 |
neurotrophin production(GO:0032898) |
| 0.1 |
0.6 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.1 |
1.7 |
GO:0046134 |
pyrimidine nucleoside biosynthetic process(GO:0046134) |
| 0.1 |
0.3 |
GO:0050884 |
neuromuscular process controlling posture(GO:0050884) |
| 0.1 |
0.4 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.1 |
0.1 |
GO:0060676 |
ureteric bud formation(GO:0060676) mesonephric tubule formation(GO:0072172) |
| 0.1 |
0.1 |
GO:0061188 |
regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 |
0.1 |
GO:0002504 |
antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 |
0.2 |
GO:0007109 |
obsolete cytokinesis, completion of separation(GO:0007109) |
| 0.1 |
0.3 |
GO:0000089 |
mitotic metaphase(GO:0000089) |
| 0.1 |
0.1 |
GO:0045005 |
DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.1 |
0.1 |
GO:0042107 |
cytokine metabolic process(GO:0042107) |
| 0.1 |
0.3 |
GO:0048630 |
skeletal muscle tissue growth(GO:0048630) |
| 0.1 |
0.3 |
GO:0003070 |
age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) |
| 0.1 |
0.2 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.1 |
0.4 |
GO:0002726 |
positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 |
0.4 |
GO:0060048 |
cardiac muscle contraction(GO:0060048) |
| 0.1 |
0.1 |
GO:0070050 |
neuron cellular homeostasis(GO:0070050) |
| 0.1 |
0.3 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 |
0.5 |
GO:0042178 |
xenobiotic catabolic process(GO:0042178) |
| 0.1 |
0.3 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 |
0.4 |
GO:0006474 |
N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 |
0.3 |
GO:0014050 |
negative regulation of glutamate secretion(GO:0014050) |
| 0.1 |
0.1 |
GO:0098927 |
early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 |
0.1 |
GO:0010761 |
fibroblast migration(GO:0010761) |
| 0.1 |
0.3 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 |
0.2 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.1 |
0.4 |
GO:0045909 |
positive regulation of vasodilation(GO:0045909) |
| 0.1 |
0.3 |
GO:0060326 |
cell chemotaxis(GO:0060326) |
| 0.1 |
0.2 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.1 |
0.1 |
GO:0031646 |
positive regulation of myelination(GO:0031643) positive regulation of neurological system process(GO:0031646) |
| 0.1 |
0.1 |
GO:0007182 |
common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 |
0.4 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.1 |
0.1 |
GO:0000239 |
pachytene(GO:0000239) |
| 0.1 |
0.5 |
GO:0070189 |
kynurenine metabolic process(GO:0070189) |
| 0.1 |
0.3 |
GO:0030201 |
heparan sulfate proteoglycan biosynthetic process(GO:0015012) heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.1 |
0.1 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 |
0.4 |
GO:0071692 |
sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 |
0.1 |
GO:0090009 |
primitive streak formation(GO:0090009) |
| 0.1 |
0.2 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 |
0.1 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.1 |
0.3 |
GO:0042471 |
ear morphogenesis(GO:0042471) inner ear morphogenesis(GO:0042472) |
| 0.1 |
0.5 |
GO:0032464 |
positive regulation of protein oligomerization(GO:0032461) positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 |
0.3 |
GO:0000710 |
meiotic mismatch repair(GO:0000710) |
| 0.1 |
0.1 |
GO:1901019 |
regulation of calcium ion transmembrane transporter activity(GO:1901019) regulation of cation channel activity(GO:2001257) |
| 0.1 |
0.6 |
GO:0051319 |
mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.1 |
1.1 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.1 |
0.6 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 |
0.2 |
GO:0034629 |
cellular protein complex localization(GO:0034629) |
| 0.1 |
0.2 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 |
0.2 |
GO:0006595 |
polyamine metabolic process(GO:0006595) |
| 0.1 |
0.2 |
GO:0006391 |
transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 |
0.3 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 |
0.8 |
GO:0006044 |
N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 |
0.5 |
GO:0000076 |
DNA replication checkpoint(GO:0000076) |
| 0.1 |
0.3 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 |
0.2 |
GO:1901970 |
positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of chromosome segregation(GO:0051984) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 |
0.3 |
GO:0060766 |
negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.1 |
0.8 |
GO:0016574 |
histone ubiquitination(GO:0016574) |
| 0.1 |
0.2 |
GO:0010972 |
negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.1 |
1.8 |
GO:0000725 |
double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.1 |
0.3 |
GO:0001554 |
luteolysis(GO:0001554) |
| 0.1 |
0.1 |
GO:0032231 |
regulation of actin filament bundle assembly(GO:0032231) positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 |
0.2 |
GO:0090267 |
positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 |
0.3 |
GO:0032570 |
response to progesterone(GO:0032570) |
| 0.1 |
0.3 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 |
0.1 |
GO:0010470 |
regulation of gastrulation(GO:0010470) |
| 0.1 |
0.2 |
GO:0006368 |
transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 |
1.4 |
GO:0034199 |
activation of protein kinase A activity(GO:0034199) |
| 0.0 |
0.8 |
GO:0042776 |
mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 |
0.1 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.0 |
0.1 |
GO:0001552 |
ovarian follicle atresia(GO:0001552) |
| 0.0 |
0.1 |
GO:0043116 |
negative regulation of vascular permeability(GO:0043116) |
| 0.0 |
0.2 |
GO:0007512 |
adult heart development(GO:0007512) |
| 0.0 |
0.1 |
GO:0072334 |
UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) nucleotide transmembrane transport(GO:1901679) |
| 0.0 |
0.1 |
GO:1901142 |
insulin processing(GO:0030070) insulin metabolic process(GO:1901142) |
| 0.0 |
0.0 |
GO:2000107 |
negative regulation of lymphocyte apoptotic process(GO:0070229) negative regulation of T cell apoptotic process(GO:0070233) negative regulation of thymocyte apoptotic process(GO:0070244) negative regulation of leukocyte apoptotic process(GO:2000107) |
| 0.0 |
0.0 |
GO:0043247 |
telomere capping(GO:0016233) protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 |
0.1 |
GO:0060744 |
thelarche(GO:0042695) development of secondary female sexual characteristics(GO:0046543) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 |
0.5 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.9 |
GO:0000380 |
alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 |
0.0 |
GO:0031622 |
regulation of fever generation(GO:0031620) positive regulation of fever generation(GO:0031622) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) |
| 0.0 |
0.1 |
GO:0006663 |
platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 |
0.7 |
GO:0007204 |
positive regulation of cytosolic calcium ion concentration(GO:0007204) |
| 0.0 |
0.0 |
GO:0051937 |
catecholamine secretion(GO:0050432) catecholamine transport(GO:0051937) |
| 0.0 |
0.2 |
GO:0017055 |
base-excision repair, DNA ligation(GO:0006288) negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 |
1.7 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 |
0.1 |
GO:0048703 |
embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 |
0.3 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
2.9 |
GO:0006826 |
iron ion transport(GO:0006826) |
| 0.0 |
0.1 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.0 |
0.1 |
GO:0007028 |
cytoplasm organization(GO:0007028) |
| 0.0 |
0.0 |
GO:0003382 |
epithelial cell morphogenesis(GO:0003382) |
| 0.0 |
0.6 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.3 |
GO:0050912 |
detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 |
0.9 |
GO:0043486 |
histone exchange(GO:0043486) |
| 0.0 |
0.3 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 |
1.0 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.0 |
0.7 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.2 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 |
1.4 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.0 |
GO:0021895 |
cerebral cortex neuron differentiation(GO:0021895) |
| 0.0 |
1.1 |
GO:0010633 |
negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 |
0.2 |
GO:0046685 |
response to arsenic-containing substance(GO:0046685) |
| 0.0 |
0.1 |
GO:0070734 |
histone H3-K27 methylation(GO:0070734) |
| 0.0 |
0.3 |
GO:1901992 |
positive regulation of cell cycle phase transition(GO:1901989) positive regulation of mitotic cell cycle phase transition(GO:1901992) |
| 0.0 |
0.1 |
GO:0071542 |
dopaminergic neuron differentiation(GO:0071542) |
| 0.0 |
0.1 |
GO:0046322 |
negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 |
0.0 |
GO:0010891 |
negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 |
0.2 |
GO:0070509 |
calcium ion import(GO:0070509) |
| 0.0 |
0.2 |
GO:0030239 |
myofibril assembly(GO:0030239) |
| 0.0 |
1.0 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.1 |
GO:0001919 |
regulation of receptor recycling(GO:0001919) |
| 0.0 |
0.9 |
GO:0007131 |
reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 |
0.2 |
GO:2000831 |
renin-angiotensin regulation of aldosterone production(GO:0002018) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) obsolete regulation of natriuresis(GO:0003078) regulation of NAD(P)H oxidase activity(GO:0033860) positive regulation of NAD(P)H oxidase activity(GO:0033864) steroid hormone secretion(GO:0035929) corticosteroid hormone secretion(GO:0035930) mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of steroid hormone secretion(GO:2000831) regulation of corticosteroid hormone secretion(GO:2000846) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.0 |
0.2 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.0 |
GO:0032528 |
microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 |
0.2 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 |
0.2 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.0 |
0.2 |
GO:0090400 |
stress-induced premature senescence(GO:0090400) |
| 0.0 |
0.1 |
GO:0008063 |
Toll signaling pathway(GO:0008063) |
| 0.0 |
0.1 |
GO:0033590 |
response to cobalamin(GO:0033590) |
| 0.0 |
0.2 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.0 |
2.8 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.1 |
GO:0016265 |
obsolete death(GO:0016265) |
| 0.0 |
0.0 |
GO:0021859 |
pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 |
0.1 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.0 |
0.2 |
GO:0031427 |
response to methotrexate(GO:0031427) |
| 0.0 |
0.4 |
GO:0019722 |
calcium-mediated signaling(GO:0019722) |
| 0.0 |
0.1 |
GO:0051645 |
Golgi localization(GO:0051645) |
| 0.0 |
0.5 |
GO:0030511 |
positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 |
0.1 |
GO:0051290 |
protein heterotetramerization(GO:0051290) |
| 0.0 |
0.3 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.0 |
0.2 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
2.9 |
GO:0006094 |
gluconeogenesis(GO:0006094) |
| 0.0 |
0.2 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 |
0.2 |
GO:0048015 |
phosphatidylinositol-mediated signaling(GO:0048015) inositol lipid-mediated signaling(GO:0048017) |
| 0.0 |
0.3 |
GO:0007223 |
Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 |
1.8 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 |
0.4 |
GO:0043496 |
regulation of protein homodimerization activity(GO:0043496) |
| 0.0 |
0.5 |
GO:0009060 |
aerobic respiration(GO:0009060) |
| 0.0 |
0.4 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
0.1 |
GO:0070407 |
oxidation-dependent protein catabolic process(GO:0070407) |
| 0.0 |
0.2 |
GO:0032964 |
collagen biosynthetic process(GO:0032964) |
| 0.0 |
0.2 |
GO:0045916 |
negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 |
0.8 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) |
| 0.0 |
0.1 |
GO:0032776 |
DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 |
0.0 |
GO:0051790 |
short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 |
4.1 |
GO:0006333 |
chromatin assembly or disassembly(GO:0006333) |
| 0.0 |
0.0 |
GO:0060251 |
regulation of glial cell proliferation(GO:0060251) negative regulation of glial cell proliferation(GO:0060253) |
| 0.0 |
0.1 |
GO:0071474 |
cellular hyperosmotic response(GO:0071474) |
| 0.0 |
0.1 |
GO:0009301 |
snRNA transcription(GO:0009301) |
| 0.0 |
0.0 |
GO:0033033 |
negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 |
0.1 |
GO:0033262 |
obsolete telomere maintenance via telomere shortening(GO:0010834) regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 |
6.8 |
GO:0007517 |
muscle organ development(GO:0007517) |
| 0.0 |
0.8 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
1.4 |
GO:0043547 |
positive regulation of GTPase activity(GO:0043547) |
| 0.0 |
0.1 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 |
0.1 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 |
0.6 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.3 |
GO:0090329 |
regulation of DNA-dependent DNA replication(GO:0090329) |
| 0.0 |
0.1 |
GO:0043278 |
response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 |
0.3 |
GO:0010507 |
negative regulation of autophagy(GO:0010507) |
| 0.0 |
0.4 |
GO:0032784 |
regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 |
0.6 |
GO:0045669 |
positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 |
0.1 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.2 |
GO:0016561 |
protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 |
0.3 |
GO:0030252 |
growth hormone secretion(GO:0030252) |
| 0.0 |
0.3 |
GO:0030422 |
production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 |
0.2 |
GO:0006349 |
regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 |
1.0 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.1 |
GO:0050830 |
defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 |
0.1 |
GO:0048485 |
sympathetic nervous system development(GO:0048485) |
| 0.0 |
0.1 |
GO:0007090 |
obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 |
0.0 |
GO:0045161 |
neuronal ion channel clustering(GO:0045161) |
| 0.0 |
0.1 |
GO:0032713 |
negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 |
0.5 |
GO:0043101 |
purine-containing compound salvage(GO:0043101) |
| 0.0 |
0.1 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.0 |
0.3 |
GO:0032332 |
positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 |
0.1 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 |
0.2 |
GO:0035455 |
response to interferon-alpha(GO:0035455) |
| 0.0 |
0.2 |
GO:0021955 |
central nervous system projection neuron axonogenesis(GO:0021952) central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 |
0.2 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.0 |
0.3 |
GO:0006700 |
C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 |
0.4 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
0.1 |
GO:0090103 |
cochlea morphogenesis(GO:0090103) |
| 0.0 |
0.5 |
GO:0001570 |
vasculogenesis(GO:0001570) |
| 0.0 |
0.1 |
GO:0051438 |
regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 |
0.2 |
GO:0034284 |
response to hexose(GO:0009746) response to monosaccharide(GO:0034284) |
| 0.0 |
0.2 |
GO:0009113 |
purine nucleobase biosynthetic process(GO:0009113) nucleobase biosynthetic process(GO:0046112) |
| 0.0 |
0.2 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 |
2.6 |
GO:0050658 |
nucleic acid transport(GO:0050657) RNA transport(GO:0050658) establishment of RNA localization(GO:0051236) |
| 0.0 |
0.1 |
GO:0033145 |
positive regulation of intracellular steroid hormone receptor signaling pathway(GO:0033145) |
| 0.0 |
0.1 |
GO:0051602 |
response to electrical stimulus(GO:0051602) |
| 0.0 |
0.1 |
GO:0071354 |
cellular response to interleukin-6(GO:0071354) |
| 0.0 |
1.2 |
GO:0034101 |
erythrocyte homeostasis(GO:0034101) |
| 0.0 |
1.0 |
GO:0000186 |
activation of MAPKK activity(GO:0000186) |
| 0.0 |
0.1 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.2 |
GO:0015755 |
fructose transport(GO:0015755) |
| 0.0 |
0.1 |
GO:0046607 |
positive regulation of centrosome cycle(GO:0046607) |
| 0.0 |
0.2 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |
| 0.0 |
0.3 |
GO:0000303 |
response to superoxide(GO:0000303) |
| 0.0 |
0.1 |
GO:0003097 |
renal water transport(GO:0003097) renal water absorption(GO:0070295) |
| 0.0 |
0.5 |
GO:0051899 |
membrane depolarization(GO:0051899) |
| 0.0 |
0.1 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.0 |
0.1 |
GO:0032693 |
negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 |
0.1 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.0 |
0.1 |
GO:0043923 |
positive regulation by host of viral transcription(GO:0043923) |
| 0.0 |
0.1 |
GO:0048535 |
lymph node development(GO:0048535) |
| 0.0 |
0.2 |
GO:0042354 |
fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 |
0.1 |
GO:0032571 |
response to vitamin K(GO:0032571) |
| 0.0 |
0.1 |
GO:0018101 |
protein citrullination(GO:0018101) citrulline biosynthetic process(GO:0019240) |
| 0.0 |
0.1 |
GO:0008588 |
release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 |
0.0 |
GO:0032674 |
interleukin-5 production(GO:0032634) regulation of interleukin-5 production(GO:0032674) |
| 0.0 |
0.1 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.0 |
0.0 |
GO:0071425 |
hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 |
0.2 |
GO:0035246 |
peptidyl-arginine methylation(GO:0018216) peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 |
0.2 |
GO:0030219 |
megakaryocyte differentiation(GO:0030219) |
| 0.0 |
0.1 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.0 |
0.2 |
GO:0060271 |
cilium morphogenesis(GO:0060271) |
| 0.0 |
0.3 |
GO:0051320 |
mitotic S phase(GO:0000084) S phase(GO:0051320) |
| 0.0 |
0.2 |
GO:0045197 |
establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 |
0.1 |
GO:0042633 |
molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 |
0.0 |
GO:0050666 |
regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.0 |
0.2 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.0 |
0.3 |
GO:0007340 |
acrosome reaction(GO:0007340) |
| 0.0 |
0.1 |
GO:0036336 |
dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 |
0.2 |
GO:0007271 |
synaptic transmission, cholinergic(GO:0007271) |
| 0.0 |
0.0 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 |
0.1 |
GO:0046476 |
glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 |
0.0 |
GO:1901976 |
regulation of spindle checkpoint(GO:0090231) regulation of cell cycle checkpoint(GO:1901976) |
| 0.0 |
0.1 |
GO:0032495 |
response to muramyl dipeptide(GO:0032495) |
| 0.0 |
0.2 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.0 |
0.5 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.0 |
0.2 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 |
0.1 |
GO:0061081 |
positive regulation of macrophage cytokine production(GO:0060907) positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) |
| 0.0 |
2.1 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 |
0.1 |
GO:0051503 |
adenine nucleotide transport(GO:0051503) |
| 0.0 |
0.0 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.0 |
0.5 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
0.4 |
GO:0046655 |
folic acid metabolic process(GO:0046655) |
| 0.0 |
0.6 |
GO:0044349 |
nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) |
| 0.0 |
0.6 |
GO:0048278 |
vesicle docking(GO:0048278) |
| 0.0 |
0.3 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.0 |
0.1 |
GO:0010875 |
positive regulation of cholesterol efflux(GO:0010875) |
| 0.0 |
0.2 |
GO:0030336 |
negative regulation of cell migration(GO:0030336) negative regulation of cell motility(GO:2000146) |
| 0.0 |
0.3 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) |
| 0.0 |
0.5 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.7 |
GO:0006813 |
potassium ion transport(GO:0006813) |
| 0.0 |
0.1 |
GO:0015858 |
nucleoside transport(GO:0015858) |
| 0.0 |
0.3 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.0 |
0.5 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.0 |
0.0 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.0 |
0.1 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 0.0 |
0.1 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
0.1 |
GO:0060370 |
susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 |
0.4 |
GO:0032608 |
interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) |
| 0.0 |
0.2 |
GO:0006069 |
ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 |
0.1 |
GO:0050906 |
detection of stimulus involved in sensory perception(GO:0050906) |
| 0.0 |
0.0 |
GO:0048845 |
venous blood vessel morphogenesis(GO:0048845) |
| 0.0 |
0.1 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 |
0.2 |
GO:0070542 |
response to fatty acid(GO:0070542) |
| 0.0 |
0.6 |
GO:0006986 |
response to unfolded protein(GO:0006986) response to topologically incorrect protein(GO:0035966) |
| 0.0 |
0.2 |
GO:0032098 |
regulation of appetite(GO:0032098) |
| 0.0 |
0.0 |
GO:0001881 |
receptor recycling(GO:0001881) |
| 0.0 |
0.0 |
GO:0060836 |
lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 |
0.2 |
GO:0001938 |
positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 |
0.4 |
GO:0007597 |
blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 |
0.0 |
GO:0031047 |
gene silencing by RNA(GO:0031047) |
| 0.0 |
0.4 |
GO:0030593 |
neutrophil chemotaxis(GO:0030593) neutrophil migration(GO:1990266) |
| 0.0 |
0.2 |
GO:0002286 |
T cell activation involved in immune response(GO:0002286) |
| 0.0 |
0.1 |
GO:0007320 |
insemination(GO:0007320) |
| 0.0 |
0.1 |
GO:0044413 |
evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 |
0.0 |
GO:0010821 |
regulation of mitochondrion organization(GO:0010821) |
| 0.0 |
0.0 |
GO:0034138 |
toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.0 |
0.4 |
GO:1902583 |
multi-organism transport(GO:0044766) transport of virus(GO:0046794) intracellular transport of virus(GO:0075733) multi-organism localization(GO:1902579) multi-organism intracellular transport(GO:1902583) |
| 0.0 |
0.4 |
GO:0031532 |
actin cytoskeleton reorganization(GO:0031532) |
| 0.0 |
0.3 |
GO:0015695 |
organic cation transport(GO:0015695) |
| 0.0 |
0.1 |
GO:0042346 |
positive regulation of NF-kappaB import into nucleus(GO:0042346) |
| 0.0 |
2.8 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.0 |
0.2 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 |
0.1 |
GO:0045329 |
amino-acid betaine metabolic process(GO:0006577) amino-acid betaine biosynthetic process(GO:0006578) carnitine metabolic process(GO:0009437) carnitine biosynthetic process(GO:0045329) |
| 0.0 |
0.4 |
GO:0017158 |
regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.0 |
0.1 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 |
0.0 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.0 |
0.2 |
GO:0015992 |
proton transport(GO:0015992) |
| 0.0 |
0.1 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 |
0.1 |
GO:0072383 |
plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 |
0.4 |
GO:0009187 |
cyclic nucleotide metabolic process(GO:0009187) |
| 0.0 |
0.0 |
GO:1903321 |
negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.0 |
0.1 |
GO:0032581 |
ER-dependent peroxisome organization(GO:0032581) |
| 0.0 |
0.0 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.9 |
GO:0000956 |
nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 |
0.1 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |
| 0.0 |
0.1 |
GO:0055083 |
monovalent inorganic anion homeostasis(GO:0055083) |
| 0.0 |
0.1 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 |
0.4 |
GO:0007585 |
respiratory gaseous exchange(GO:0007585) |
| 0.0 |
0.2 |
GO:0043487 |
regulation of RNA stability(GO:0043487) |
| 0.0 |
0.1 |
GO:0051924 |
regulation of calcium ion transport(GO:0051924) |
| 0.0 |
0.2 |
GO:0007093 |
mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 |
0.7 |
GO:0034470 |
ncRNA processing(GO:0034470) |
| 0.0 |
0.2 |
GO:0051568 |
histone H3-K4 methylation(GO:0051568) |
| 0.0 |
0.0 |
GO:0061084 |
regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) negative regulation of protein folding(GO:1903333) |
| 0.0 |
0.1 |
GO:0000354 |
cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 |
0.1 |
GO:0032508 |
DNA duplex unwinding(GO:0032508) |
| 0.0 |
0.0 |
GO:0044380 |
protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 |
0.0 |
GO:0051881 |
regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 |
0.4 |
GO:2000060 |
positive regulation of ubiquitin-protein ligase activity involved in regulation of mitotic cell cycle transition(GO:0051437) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 |
0.2 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.1 |
GO:0046500 |
S-adenosylmethionine metabolic process(GO:0046500) |
| 0.0 |
0.1 |
GO:0007528 |
neuromuscular junction development(GO:0007528) |
| 0.0 |
0.1 |
GO:0070498 |
interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 |
0.0 |
GO:0032007 |
negative regulation of TOR signaling(GO:0032007) |
| 0.0 |
0.1 |
GO:0032203 |
telomere formation via telomerase(GO:0032203) |
| 0.0 |
0.1 |
GO:0043388 |
positive regulation of DNA binding(GO:0043388) |
| 0.0 |
0.0 |
GO:0046033 |
AMP metabolic process(GO:0046033) |
| 0.0 |
0.2 |
GO:0007422 |
peripheral nervous system development(GO:0007422) |
| 0.0 |
0.1 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.0 |
0.5 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
0.4 |
GO:0031032 |
actomyosin structure organization(GO:0031032) |
| 0.0 |
0.1 |
GO:0070373 |
negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 |
0.1 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.3 |
GO:0051925 |
obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 |
1.2 |
GO:0031295 |
lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 |
0.0 |
GO:0016197 |
endosomal transport(GO:0016197) |
| 0.0 |
0.1 |
GO:0000216 |
obsolete M/G1 transition of mitotic cell cycle(GO:0000216) |
| 0.0 |
0.1 |
GO:0006787 |
porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 |
0.1 |
GO:0009154 |
purine ribonucleotide catabolic process(GO:0009154) |
| 0.0 |
0.0 |
GO:0033146 |
regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 |
0.1 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.1 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.0 |
GO:0032604 |
granulocyte macrophage colony-stimulating factor production(GO:0032604) regulation of granulocyte macrophage colony-stimulating factor production(GO:0032645) positive regulation of granulocyte macrophage colony-stimulating factor production(GO:0032725) |
| 0.0 |
0.2 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.1 |
GO:0009103 |
lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 |
0.0 |
GO:0009067 |
aspartate family amino acid biosynthetic process(GO:0009067) |
| 0.0 |
0.1 |
GO:0006658 |
phosphatidylserine metabolic process(GO:0006658) |
| 0.0 |
0.1 |
GO:0071025 |
RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 |
0.1 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.2 |
GO:0043124 |
negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 |
0.0 |
GO:0045778 |
positive regulation of ossification(GO:0045778) |
| 0.0 |
0.1 |
GO:0048477 |
oogenesis(GO:0048477) |
| 0.0 |
0.1 |
GO:0018210 |
peptidyl-threonine modification(GO:0018210) |
| 0.0 |
0.1 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.0 |
0.1 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.0 |
0.0 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.0 |
0.0 |
GO:0019255 |
glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 |
0.0 |
GO:1903729 |
regulation of plasma membrane organization(GO:1903729) |
| 0.0 |
0.0 |
GO:0019348 |
polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 |
0.1 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.0 |
0.0 |
GO:0002019 |
regulation of renal output by angiotensin(GO:0002019) |
| 0.0 |
0.3 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.0 |
0.0 |
GO:0002467 |
germinal center formation(GO:0002467) |
| 0.0 |
0.1 |
GO:0022616 |
DNA strand elongation(GO:0022616) |
| 0.0 |
0.0 |
GO:0033962 |
cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 |
0.1 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.0 |
0.0 |
GO:0006999 |
nuclear pore organization(GO:0006999) |
| 0.0 |
0.2 |
GO:0042136 |
neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 |
0.0 |
GO:0043558 |
regulation of translation in response to stress(GO:0043555) regulation of translational initiation in response to stress(GO:0043558) |
| 0.0 |
5.7 |
GO:0006518 |
peptide metabolic process(GO:0006518) |
| 0.0 |
0.1 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 |
0.0 |
GO:0009791 |
post-embryonic development(GO:0009791) |
| 0.0 |
0.0 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.0 |
0.0 |
GO:0006848 |
pyruvate transport(GO:0006848) |
| 0.0 |
0.0 |
GO:0019987 |
obsolete negative regulation of anti-apoptosis(GO:0019987) |