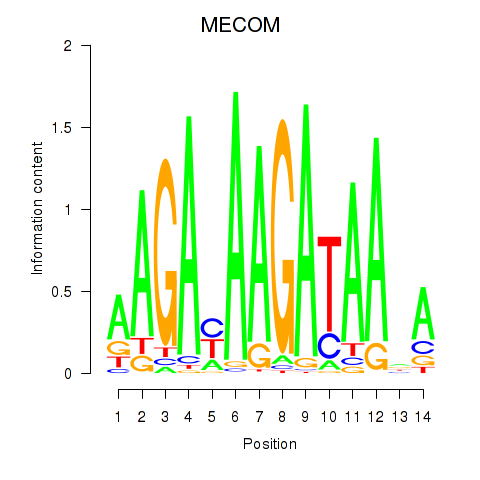
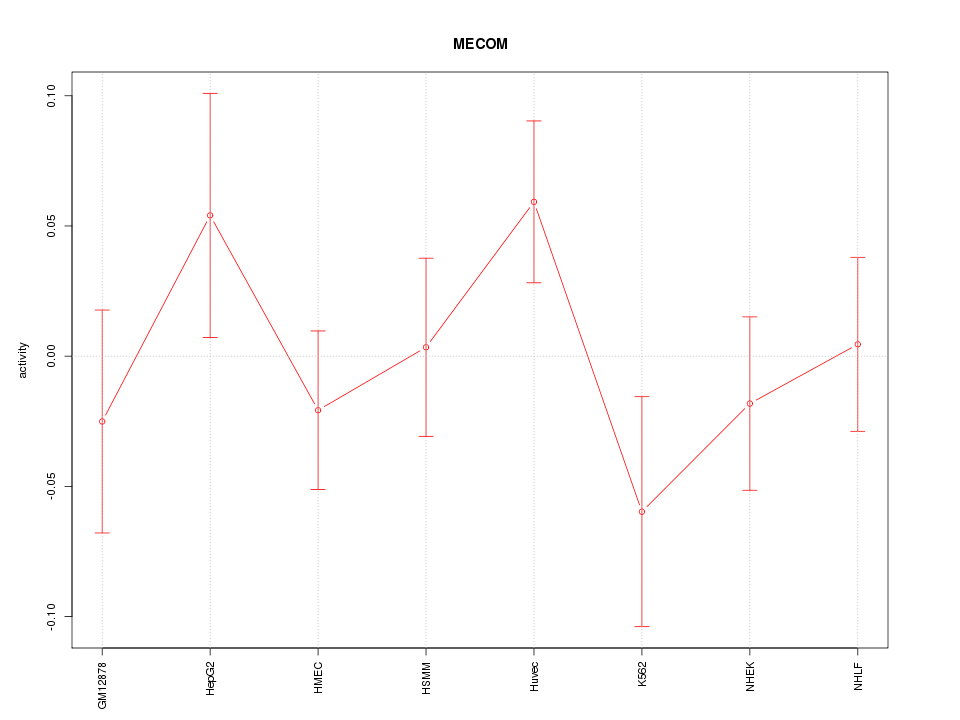
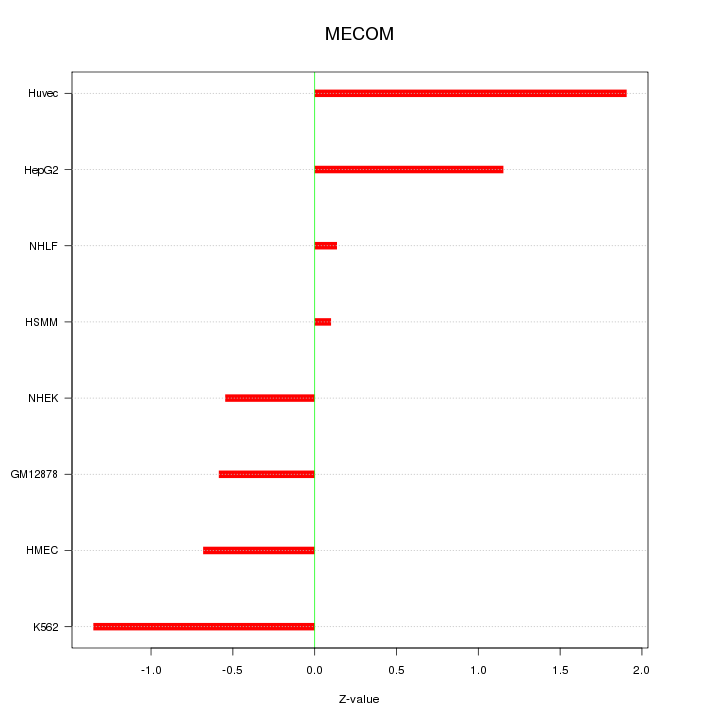
| 1.2 |
4.9 |
GO:0050928 |
negative regulation of positive chemotaxis(GO:0050928) |
| 0.7 |
2.1 |
GO:0001907 |
cytolysis by symbiont of host cells(GO:0001897) killing by symbiont of host cells(GO:0001907) hemolysis by symbiont of host erythrocytes(GO:0019836) disruption by symbiont of host cell(GO:0044004) hemolysis in other organism(GO:0044179) positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) cytolysis in other organism(GO:0051715) cytolysis in other organism involved in symbiotic interaction(GO:0051801) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.5 |
1.6 |
GO:2000910 |
regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.4 |
3.6 |
GO:0017121 |
phospholipid scrambling(GO:0017121) |
| 0.2 |
0.5 |
GO:0032488 |
Cdc42 protein signal transduction(GO:0032488) |
| 0.2 |
0.5 |
GO:0014063 |
regulation of serotonin secretion(GO:0014062) negative regulation of serotonin secretion(GO:0014063) |
| 0.1 |
1.2 |
GO:0060261 |
positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 |
1.1 |
GO:0042448 |
progesterone metabolic process(GO:0042448) |
| 0.1 |
0.4 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 |
0.6 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.1 |
0.3 |
GO:0090598 |
male genitalia morphogenesis(GO:0048808) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 |
0.4 |
GO:0033622 |
integrin activation(GO:0033622) regulation of integrin activation(GO:0033623) positive regulation of integrin activation(GO:0033625) |
| 0.1 |
0.4 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 |
0.4 |
GO:0070245 |
positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 |
1.0 |
GO:0032331 |
negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 |
0.2 |
GO:0045872 |
positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 |
0.4 |
GO:0099515 |
nuclear migration along microfilament(GO:0031022) actin filament-based transport(GO:0099515) |
| 0.1 |
1.0 |
GO:0001502 |
cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 |
0.1 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 |
0.1 |
GO:0014831 |
intestine smooth muscle contraction(GO:0014827) gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.0 |
0.1 |
GO:0034126 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) positive regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034126) |
| 0.0 |
0.1 |
GO:0043628 |
ncRNA 3'-end processing(GO:0043628) |
| 0.0 |
0.4 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.0 |
1.7 |
GO:0006749 |
glutathione metabolic process(GO:0006749) |
| 0.0 |
0.2 |
GO:1901985 |
positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 |
0.3 |
GO:0097435 |
extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.0 |
0.3 |
GO:0010875 |
positive regulation of cholesterol efflux(GO:0010875) |
| 0.0 |
0.4 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.0 |
0.3 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
0.8 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.3 |
GO:0045749 |
obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 |
0.1 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
1.1 |
GO:0001764 |
neuron migration(GO:0001764) |
| 0.0 |
0.4 |
GO:0017144 |
drug metabolic process(GO:0017144) |
| 0.0 |
0.0 |
GO:0042268 |
regulation of cytolysis(GO:0042268) |
| 0.0 |
1.1 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.0 |
0.1 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 |
0.3 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |