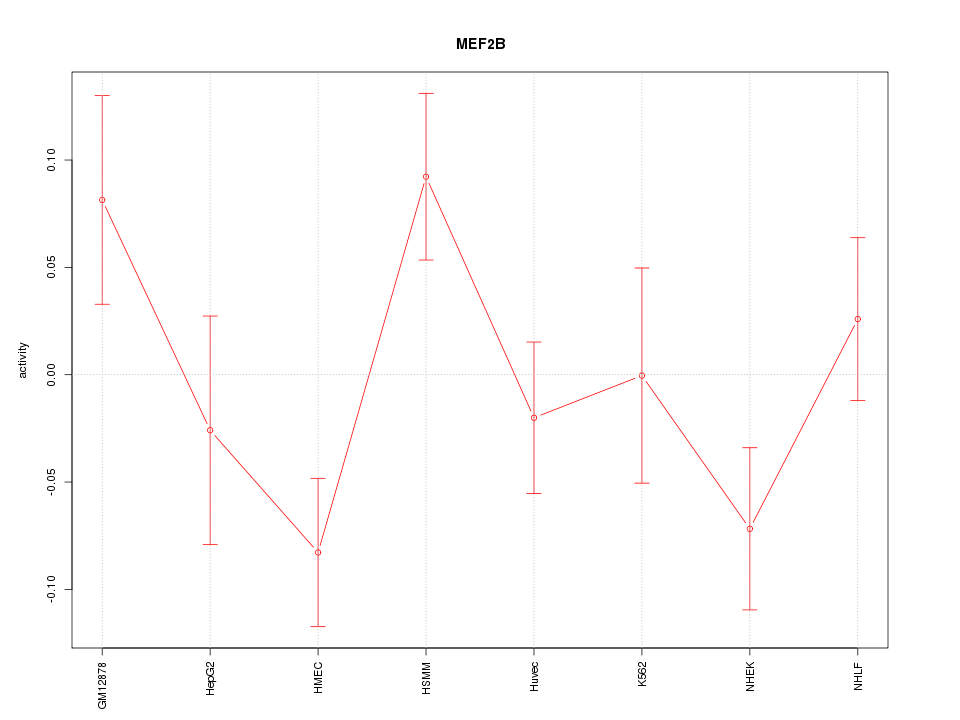
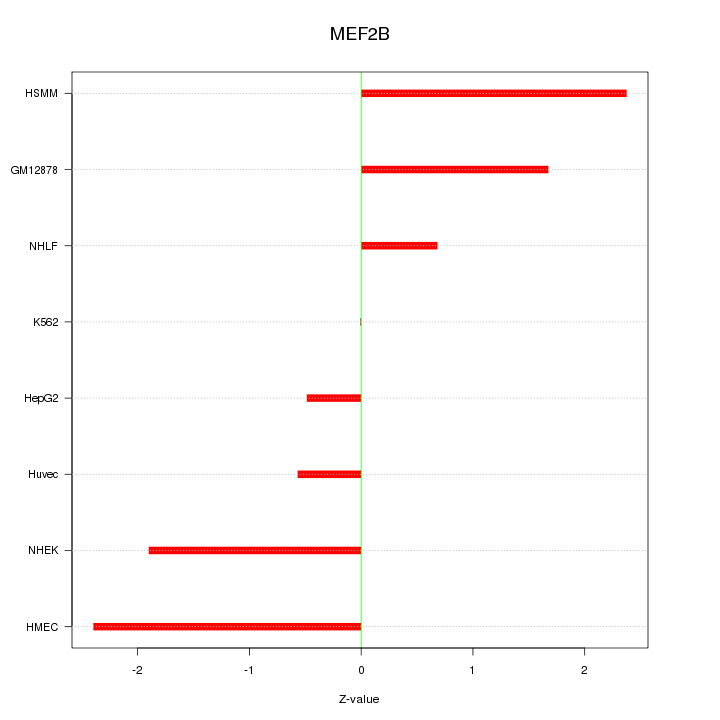
Motif ID: MEF2B
Z-value: 1.534
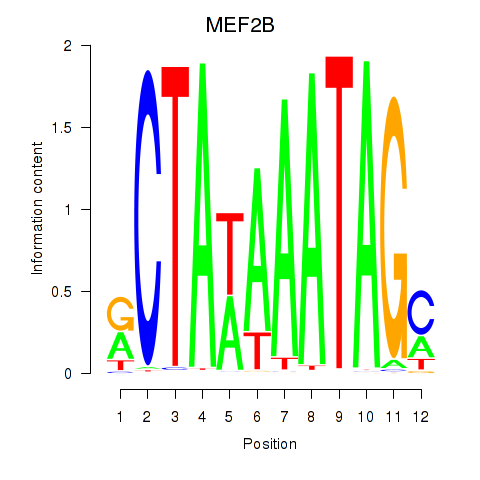
Transcription factors associated with MEF2B:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| MEF2B | ENSG00000213999.11 | MEF2B |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.6 | 2.8 | GO:0003093 | regulation of glomerular filtration(GO:0003093) glomerular filtration(GO:0003094) renal filtration(GO:0097205) regulation of renal system process(GO:0098801) |
| 0.5 | 1.6 | GO:1903115 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.2 | 2.9 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.1 | 1.5 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.1 | 1.2 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.4 | GO:0048861 | serine phosphorylation of STAT protein(GO:0042501) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 1.1 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.1 | 1.0 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 2.9 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 5.3 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 0.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 4.9 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 1.1 | GO:0045069 | regulation of viral genome replication(GO:0045069) |
| 0.0 | 0.2 | GO:0051154 | negative regulation of myotube differentiation(GO:0010832) negative regulation of striated muscle cell differentiation(GO:0051154) |
| 0.0 | 1.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 2.4 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.0 | 0.2 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 1.6 | GO:0051149 | positive regulation of muscle cell differentiation(GO:0051149) |
| 0.0 | 0.6 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.0 | 4.9 | GO:0006184 | obsolete GTP catabolic process(GO:0006184) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) epithelial fluid transport(GO:0042045) cellular response to copper ion(GO:0071280) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 1.5 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 0.2 | GO:0006091 | generation of precursor metabolites and energy(GO:0006091) |
| 0.0 | 0.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.0 | GO:0006094 | gluconeogenesis(GO:0006094) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 2.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.3 | 2.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 3.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.0 | GO:0032982 | myosin filament(GO:0032982) |
| 0.1 | 0.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 1.9 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.2 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.1 | GO:0033655 | host(GO:0018995) host cell cytoplasm(GO:0030430) host cell part(GO:0033643) host intracellular part(GO:0033646) host cell cytoplasm part(GO:0033655) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 1.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.8 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0008907 | integrase activity(GO:0008907) |
| 0.2 | 4.9 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.2 | 2.8 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 2.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.2 | 1.8 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.2 | 4.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 2.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.9 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 1.0 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 1.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 1.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.4 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 2.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 2.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 4.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.7 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.3 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.2 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |