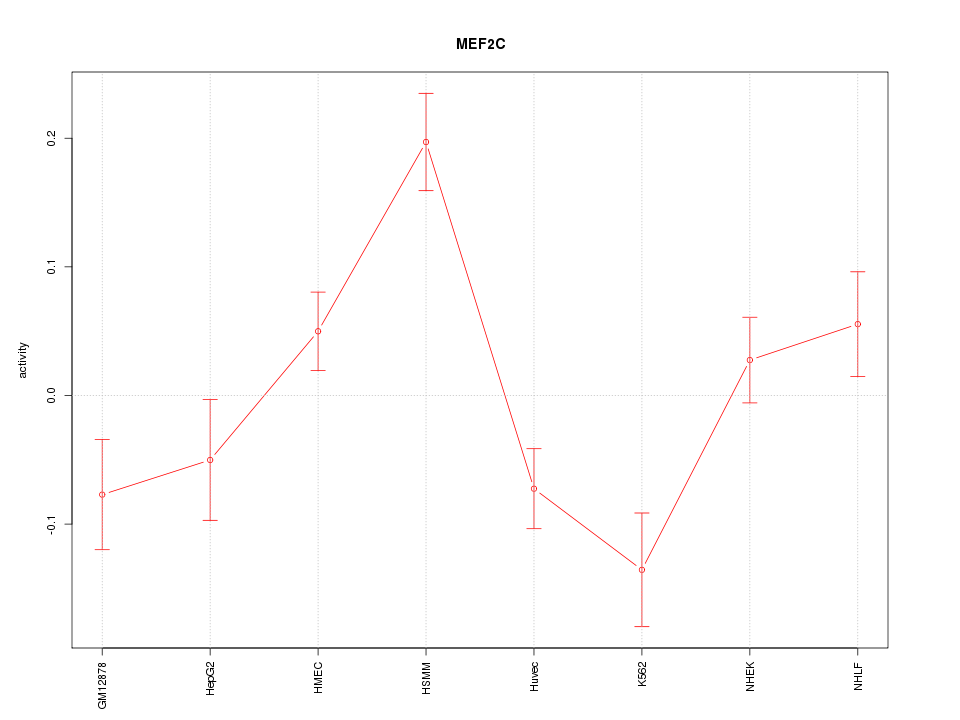
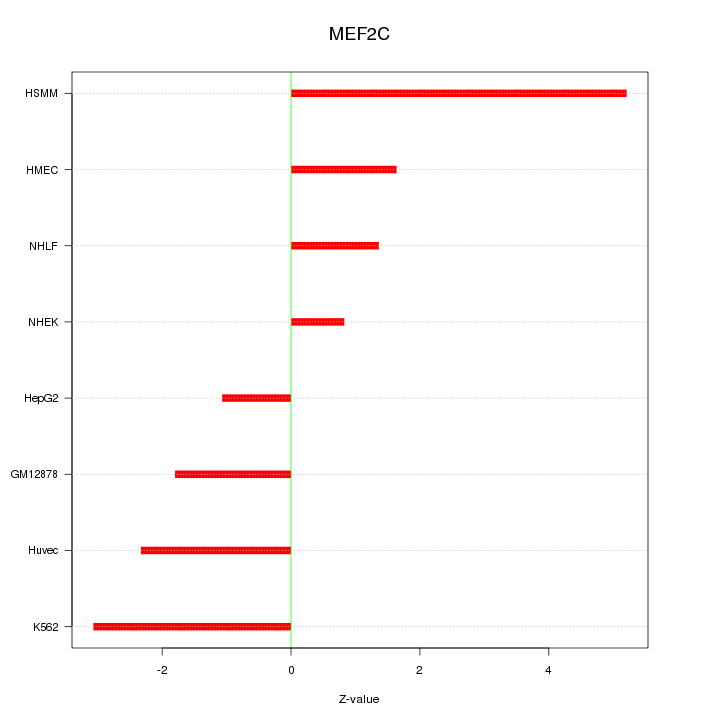
Motif ID: MEF2C
Z-value: 2.539
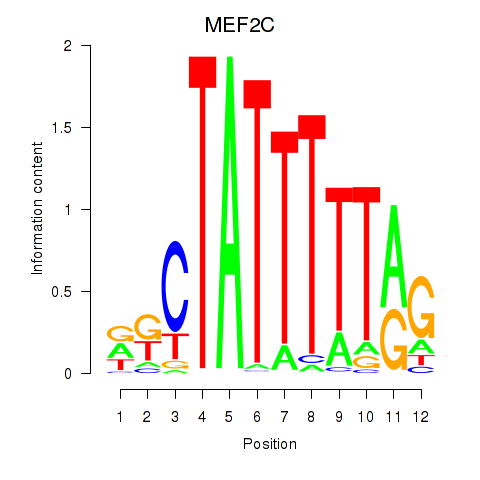
Transcription factors associated with MEF2C:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| MEF2C | ENSG00000081189.9 | MEF2C |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 2.1 | 12.7 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 1.6 | 4.8 | GO:1903115 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 1.0 | 2.9 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.6 | 5.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.6 | 3.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.6 | 20.4 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.5 | 1.6 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.4 | 2.9 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.4 | 1.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.4 | 1.9 | GO:0090343 | positive regulation of cell aging(GO:0090343) |
| 0.4 | 1.1 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.3 | 5.3 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.2 | 2.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.2 | 0.5 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 | 1.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.2 | 4.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 3.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 1.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.1 | 3.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 21.8 | GO:0006184 | obsolete GTP catabolic process(GO:0006184) |
| 0.1 | 0.3 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) response to cobalt ion(GO:0032025) |
| 0.1 | 0.5 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 2.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 2.0 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.1 | 0.3 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.6 | GO:0019934 | cGMP-mediated signaling(GO:0019934) regulation of phagocytosis, engulfment(GO:0060099) positive regulation of phagocytosis, engulfment(GO:0060100) regulation of membrane invagination(GO:1905153) positive regulation of membrane invagination(GO:1905155) |
| 0.1 | 2.1 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 5.6 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 0.7 | GO:0045408 | regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.1 | 0.7 | GO:0060438 | trachea development(GO:0060438) |
| 0.1 | 0.4 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.4 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 1.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.7 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 1.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 2.1 | GO:0051149 | positive regulation of muscle cell differentiation(GO:0051149) |
| 0.0 | 3.1 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 7.6 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.7 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 6.1 | GO:0050658 | nucleic acid transport(GO:0050657) RNA transport(GO:0050658) establishment of RNA localization(GO:0051236) |
| 0.0 | 0.8 | GO:0051925 | obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 | 0.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 3.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.4 | GO:0006090 | pyruvate metabolic process(GO:0006090) |
| 0.0 | 0.4 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) actin-mediated cell contraction(GO:0070252) |
| 0.0 | 0.7 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.5 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 1.5 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 11.7 | GO:0007010 | cytoskeleton organization(GO:0007010) |
| 0.0 | 6.6 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.4 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0048268 | negative regulation of receptor-mediated endocytosis(GO:0048261) clathrin coat assembly(GO:0048268) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 32.6 | GO:0005861 | troponin complex(GO:0005861) |
| 1.2 | 3.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 2.7 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.5 | 2.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.5 | 4.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.3 | 16.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 1.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 1.4 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 4.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 2.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.7 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.1 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 4.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.5 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 2.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 18.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 1.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 6.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.5 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 4.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 2.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.6 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.1 | GO:0035097 | methyltransferase complex(GO:0034708) histone methyltransferase complex(GO:0035097) |
| 0.0 | 2.9 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 1.9 | GO:0031965 | nuclear membrane(GO:0031965) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 32.3 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 2.7 | 16.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 1.1 | 12.0 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.8 | 3.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.5 | 10.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.5 | 2.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.5 | 5.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.5 | 1.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.4 | 8.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.4 | 2.9 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.3 | 1.5 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 2.4 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.2 | 1.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 0.7 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 2.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 4.6 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.1 | 4.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.5 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.6 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 1.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 2.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.5 | GO:0004528 | phosphodiesterase I activity(GO:0004528) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.1 | 2.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 5.4 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 17.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 1.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 1.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 0.2 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 1.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 6.7 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 8.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 3.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 6.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 10.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 2.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 7.0 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 16.2 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.1 | 1.6 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.7 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |