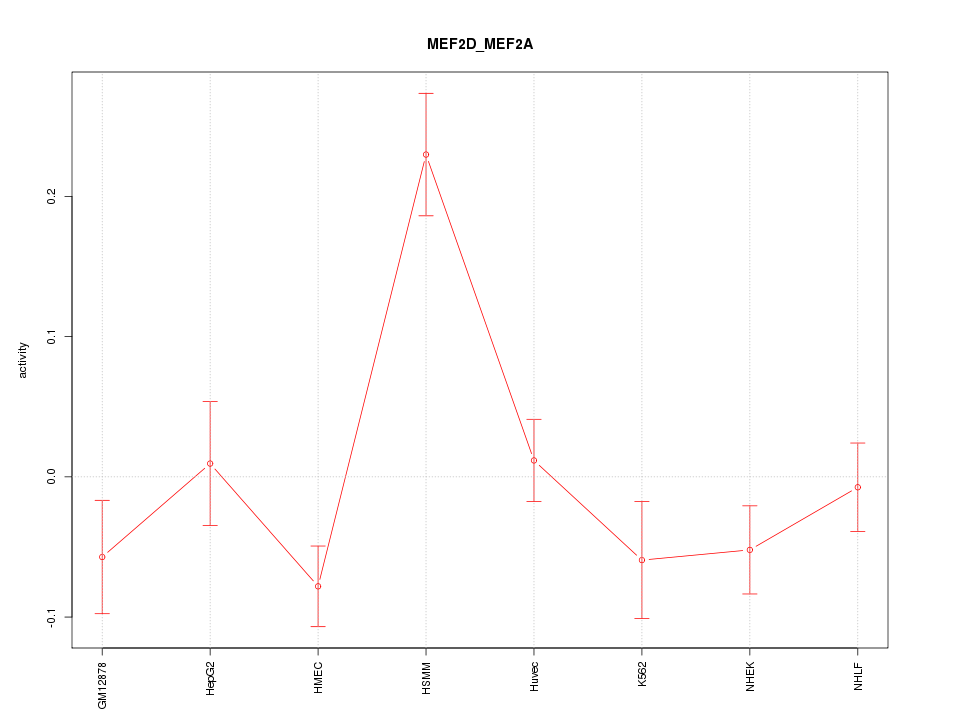
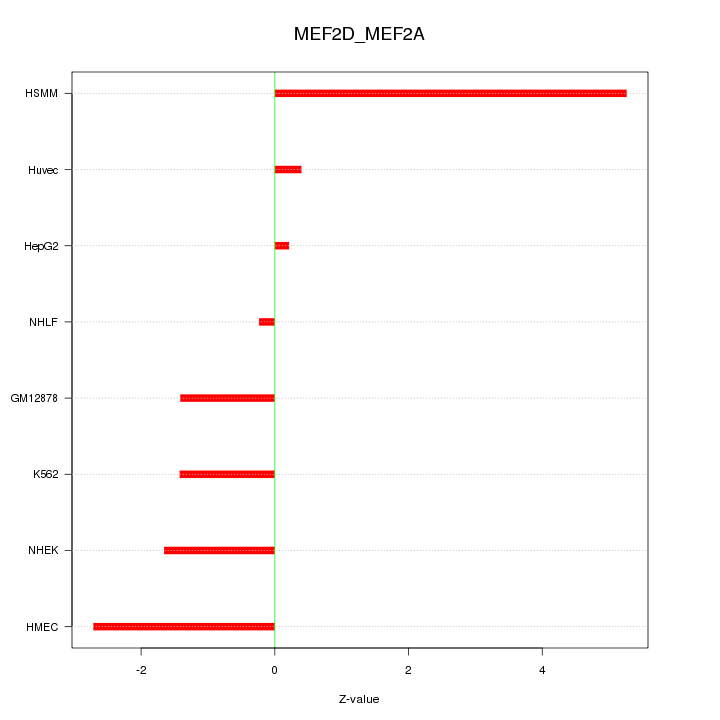
Motif ID: MEF2D_MEF2A
Z-value: 2.294
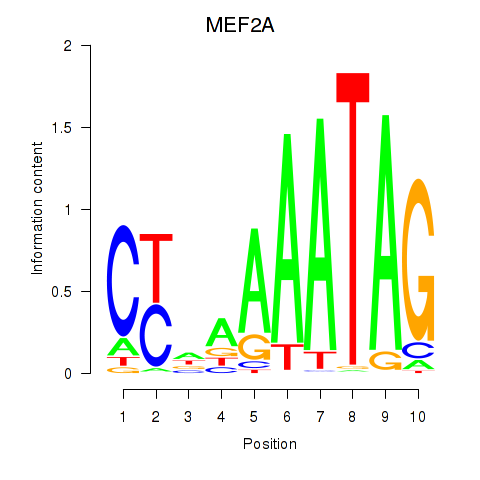
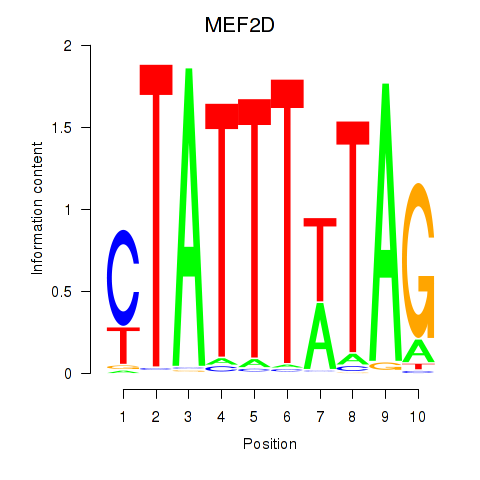
Transcription factors associated with MEF2D_MEF2A:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| MEF2A | ENSG00000068305.13 | MEF2A |
| MEF2D | ENSG00000116604.13 | MEF2D |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:1903115 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 1.7 | 5.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 1.3 | 3.8 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.8 | 27.3 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.7 | 3.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.7 | 2.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.6 | 3.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.5 | 12.1 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.4 | 1.5 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.4 | 3.2 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.3 | 1.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.3 | 13.8 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.3 | 1.6 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.2 | 2.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 0.6 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.2 | 5.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 1.0 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.2 | 2.1 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 0.7 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.8 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 4.2 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.1 | 0.7 | GO:0090343 | positive regulation of cell aging(GO:0090343) |
| 0.1 | 0.5 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 10.9 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 4.3 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 0.3 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 1.8 | GO:0042749 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 24.1 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.1 | 0.6 | GO:0015811 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.1 | 0.8 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.8 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.1 | 1.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 1.8 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.5 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) behavioral response to cocaine(GO:0048148) |
| 0.1 | 0.6 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.1 | 1.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 2.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 4.0 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 3.6 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.1 | 0.5 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 1.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.5 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 1.2 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.4 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.9 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 1.0 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.4 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 5.8 | GO:0050658 | nucleic acid transport(GO:0050657) RNA transport(GO:0050658) establishment of RNA localization(GO:0051236) |
| 0.0 | 0.2 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 2.5 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.4 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.8 | GO:0051925 | obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 | 0.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 1.5 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 2.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.0 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 2.0 | GO:0036293 | response to hypoxia(GO:0001666) response to decreased oxygen levels(GO:0036293) |
| 0.0 | 1.8 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.5 | 35.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.8 | 8.0 | GO:0031430 | M band(GO:0031430) |
| 0.7 | 17.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.6 | 4.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.5 | 3.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.5 | 2.5 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.5 | 4.4 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.3 | 2.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 2.5 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.3 | 3.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 2.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 1.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 3.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 6.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.5 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 2.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 0.7 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.1 | 6.1 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 0.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.4 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.1 | 1.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 5.4 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 1.6 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 2.5 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 4.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 4.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 5.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 3.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 2.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 14.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 1.0 | 3.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.9 | 18.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.8 | 4.1 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.6 | 3.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.6 | 14.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.5 | 3.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.5 | 2.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.5 | 3.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 1.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.4 | 3.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.4 | 1.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.4 | 2.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.3 | 2.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 0.8 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.3 | 23.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 3.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 0.6 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.2 | 2.4 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.2 | 0.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 1.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 3.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 1.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.5 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 3.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 3.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 3.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 0.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 6.9 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 2.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 5.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.5 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 1.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.4 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0015254 | urea transmembrane transporter activity(GO:0015204) glycerol channel activity(GO:0015254) |
| 0.0 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 12.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 2.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.9 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 2.3 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 4.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.0 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 9.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 2.2 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 2.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 19.7 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.1 | 2.4 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.0 | 1.0 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | SIG_PIP3_SIGNALING_IN_B_LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |