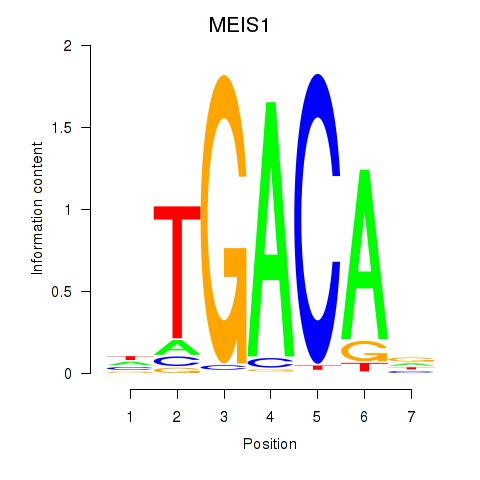
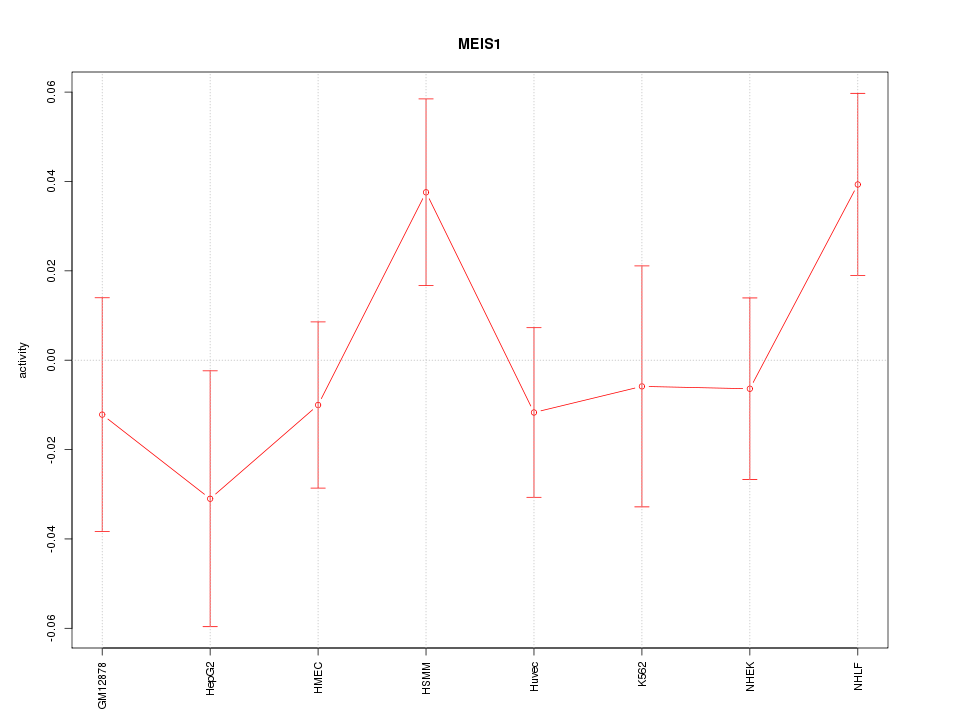
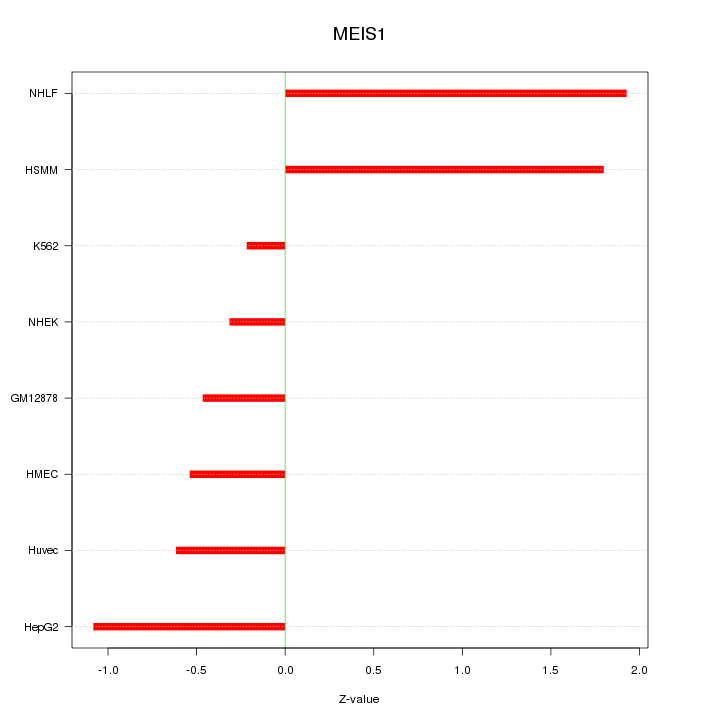
| 0.4 |
0.4 |
GO:0016573 |
internal protein amino acid acetylation(GO:0006475) histone acetylation(GO:0016573) internal peptidyl-lysine acetylation(GO:0018393) peptidyl-lysine acetylation(GO:0018394) |
| 0.4 |
1.5 |
GO:0002689 |
negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.3 |
1.0 |
GO:1903115 |
regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.3 |
1.2 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.3 |
1.7 |
GO:2000124 |
cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 |
0.6 |
GO:0060028 |
convergent extension involved in axis elongation(GO:0060028) |
| 0.3 |
0.8 |
GO:0031034 |
skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 |
0.8 |
GO:2000121 |
regulation of removal of superoxide radicals(GO:2000121) |
| 0.3 |
2.0 |
GO:0007185 |
transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.3 |
3.5 |
GO:1901797 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.2 |
0.2 |
GO:0003350 |
pulmonary myocardium development(GO:0003350) |
| 0.2 |
1.4 |
GO:0007499 |
ectoderm and mesoderm interaction(GO:0007499) |
| 0.2 |
0.9 |
GO:0043116 |
negative regulation of vascular permeability(GO:0043116) |
| 0.2 |
0.4 |
GO:0048319 |
axial mesoderm morphogenesis(GO:0048319) |
| 0.2 |
0.7 |
GO:0021569 |
rhombomere 3 development(GO:0021569) |
| 0.2 |
1.5 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 |
1.4 |
GO:0071692 |
sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 |
0.3 |
GO:0090598 |
male genitalia morphogenesis(GO:0048808) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 |
0.4 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.1 |
0.9 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 |
0.4 |
GO:0050968 |
detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 |
0.3 |
GO:0046931 |
pore complex assembly(GO:0046931) |
| 0.1 |
0.3 |
GO:0032242 |
regulation of nucleoside transport(GO:0032242) regulation of neurotrophin production(GO:0032899) negative regulation of neurotrophin production(GO:0032900) negative regulation of vasodilation(GO:0045908) |
| 0.1 |
0.3 |
GO:0046586 |
regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 |
0.5 |
GO:0014819 |
regulation of skeletal muscle contraction(GO:0014819) |
| 0.1 |
0.5 |
GO:0044413 |
evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 |
1.2 |
GO:0032331 |
negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 |
0.4 |
GO:0008354 |
germ cell migration(GO:0008354) |
| 0.1 |
2.0 |
GO:0008347 |
glial cell migration(GO:0008347) |
| 0.1 |
1.7 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.1 |
0.3 |
GO:0042262 |
DNA protection(GO:0042262) |
| 0.1 |
0.4 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 |
1.8 |
GO:0042730 |
fibrinolysis(GO:0042730) |
| 0.1 |
0.2 |
GO:0050758 |
thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 |
0.2 |
GO:0006740 |
NADPH regeneration(GO:0006740) |
| 0.1 |
0.4 |
GO:0022614 |
membrane to membrane docking(GO:0022614) chorio-allantoic fusion(GO:0060710) |
| 0.1 |
0.7 |
GO:0048261 |
negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.1 |
5.0 |
GO:0009408 |
response to heat(GO:0009408) |
| 0.1 |
0.8 |
GO:0060638 |
mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.1 |
0.3 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
0.2 |
GO:2000858 |
renin-angiotensin regulation of aldosterone production(GO:0002018) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) positive regulation of NAD(P)H oxidase activity(GO:0033864) mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.1 |
0.1 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
0.3 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.1 |
0.2 |
GO:0033364 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mast cell secretory granule organization(GO:0033364) |
| 0.1 |
0.1 |
GO:0032509 |
endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 |
0.1 |
GO:0070781 |
response to biotin(GO:0070781) |
| 0.0 |
0.1 |
GO:1903960 |
negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 |
1.6 |
GO:0060048 |
cardiac muscle contraction(GO:0060048) |
| 0.0 |
0.5 |
GO:0006942 |
regulation of striated muscle contraction(GO:0006942) |
| 0.0 |
0.4 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |
| 0.0 |
0.3 |
GO:0032801 |
receptor catabolic process(GO:0032801) |
| 0.0 |
0.5 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
0.1 |
GO:0014002 |
astrocyte development(GO:0014002) |
| 0.0 |
0.8 |
GO:0032012 |
regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.4 |
GO:0009263 |
deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 |
0.4 |
GO:0045932 |
negative regulation of muscle contraction(GO:0045932) |
| 0.0 |
0.2 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.2 |
GO:0043001 |
Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 |
0.2 |
GO:0006295 |
nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 |
2.3 |
GO:0006892 |
post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 |
0.4 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.0 |
0.1 |
GO:1902913 |
positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.0 |
0.5 |
GO:0015939 |
pantothenate metabolic process(GO:0015939) |
| 0.0 |
0.2 |
GO:0032236 |
obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 |
1.1 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 |
0.2 |
GO:0035455 |
response to interferon-alpha(GO:0035455) |
| 0.0 |
0.1 |
GO:0043587 |
tongue morphogenesis(GO:0043587) |
| 0.0 |
0.1 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.0 |
0.1 |
GO:0046520 |
diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.0 |
0.1 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 0.0 |
0.4 |
GO:0051496 |
positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 |
0.1 |
GO:0031558 |
obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 |
0.0 |
GO:0003078 |
obsolete regulation of natriuresis(GO:0003078) |
| 0.0 |
0.2 |
GO:0019934 |
cGMP-mediated signaling(GO:0019934) |
| 0.0 |
0.5 |
GO:0044349 |
nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) |
| 0.0 |
0.4 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.0 |
0.3 |
GO:0006600 |
creatine metabolic process(GO:0006600) |
| 0.0 |
0.2 |
GO:0032472 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.0 |
0.1 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 |
0.1 |
GO:0090047 |
retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 |
0.8 |
GO:0001755 |
neural crest cell migration(GO:0001755) |
| 0.0 |
0.1 |
GO:0060831 |
smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 |
0.2 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.0 |
0.1 |
GO:0044321 |
cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 |
0.2 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.0 |
0.0 |
GO:0010834 |
obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 |
0.8 |
GO:0050690 |
regulation of defense response to virus by virus(GO:0050690) |
| 0.0 |
1.2 |
GO:0051318 |
G1 phase(GO:0051318) |
| 0.0 |
0.1 |
GO:0022417 |
protein maturation by protein folding(GO:0022417) |
| 0.0 |
0.1 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.4 |
GO:0000303 |
response to superoxide(GO:0000303) |
| 0.0 |
0.1 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.0 |
GO:0007632 |
visual behavior(GO:0007632) |
| 0.0 |
0.1 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 |
0.8 |
GO:0009060 |
aerobic respiration(GO:0009060) |
| 0.0 |
0.1 |
GO:0000212 |
meiotic spindle organization(GO:0000212) |
| 0.0 |
0.1 |
GO:0001514 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.3 |
GO:0031000 |
response to caffeine(GO:0031000) |
| 0.0 |
0.4 |
GO:0043496 |
regulation of protein homodimerization activity(GO:0043496) |
| 0.0 |
0.1 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 |
0.0 |
GO:0002281 |
macrophage activation involved in immune response(GO:0002281) |
| 0.0 |
0.1 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 |
0.2 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 |
0.2 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
2.2 |
GO:0065004 |
protein-DNA complex assembly(GO:0065004) |
| 0.0 |
0.1 |
GO:0032781 |
positive regulation of ATPase activity(GO:0032781) |
| 0.0 |
0.1 |
GO:0005984 |
disaccharide metabolic process(GO:0005984) |
| 0.0 |
1.1 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
0.1 |
GO:0042074 |
cell migration involved in gastrulation(GO:0042074) |
| 0.0 |
0.2 |
GO:0009148 |
GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine nucleoside triphosphate biosynthetic process(GO:0009148) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.0 |
0.5 |
GO:0042771 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 |
0.2 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.0 |
0.2 |
GO:0051927 |
obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 |
0.5 |
GO:0007368 |
determination of left/right symmetry(GO:0007368) |
| 0.0 |
1.0 |
GO:0006826 |
iron ion transport(GO:0006826) |
| 0.0 |
0.1 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.0 |
GO:0035269 |
protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 |
0.6 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.5 |
GO:0007566 |
embryo implantation(GO:0007566) |
| 0.0 |
0.1 |
GO:0042483 |
negative regulation of odontogenesis(GO:0042483) |
| 0.0 |
0.2 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) |
| 0.0 |
0.0 |
GO:0046469 |
platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 |
0.2 |
GO:0033138 |
positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 |
0.2 |
GO:0030166 |
proteoglycan biosynthetic process(GO:0030166) |
| 0.0 |
0.2 |
GO:0008634 |
obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 |
0.1 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.2 |
GO:1904031 |
positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 |
0.1 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.1 |
GO:0006216 |
cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 |
0.2 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 |
0.3 |
GO:0006084 |
acetyl-CoA metabolic process(GO:0006084) |
| 0.0 |
0.1 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.1 |
GO:0031572 |
G2 DNA damage checkpoint(GO:0031572) |
| 0.0 |
0.1 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.2 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
1.5 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.1 |
GO:0045329 |
amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 |
2.6 |
GO:0006936 |
muscle contraction(GO:0006936) |
| 0.0 |
0.1 |
GO:0000710 |
meiotic mismatch repair(GO:0000710) |
| 0.0 |
0.1 |
GO:0006196 |
AMP catabolic process(GO:0006196) |
| 0.0 |
0.3 |
GO:0005978 |
glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 |
0.1 |
GO:0061756 |
leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 |
0.1 |
GO:0042178 |
xenobiotic catabolic process(GO:0042178) |
| 0.0 |
0.0 |
GO:0051102 |
DNA ligation involved in DNA recombination(GO:0051102) |