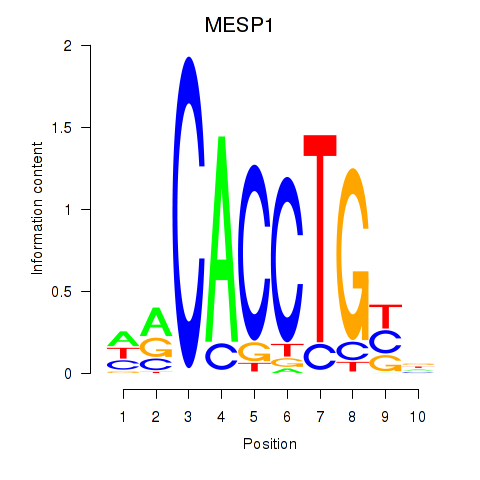
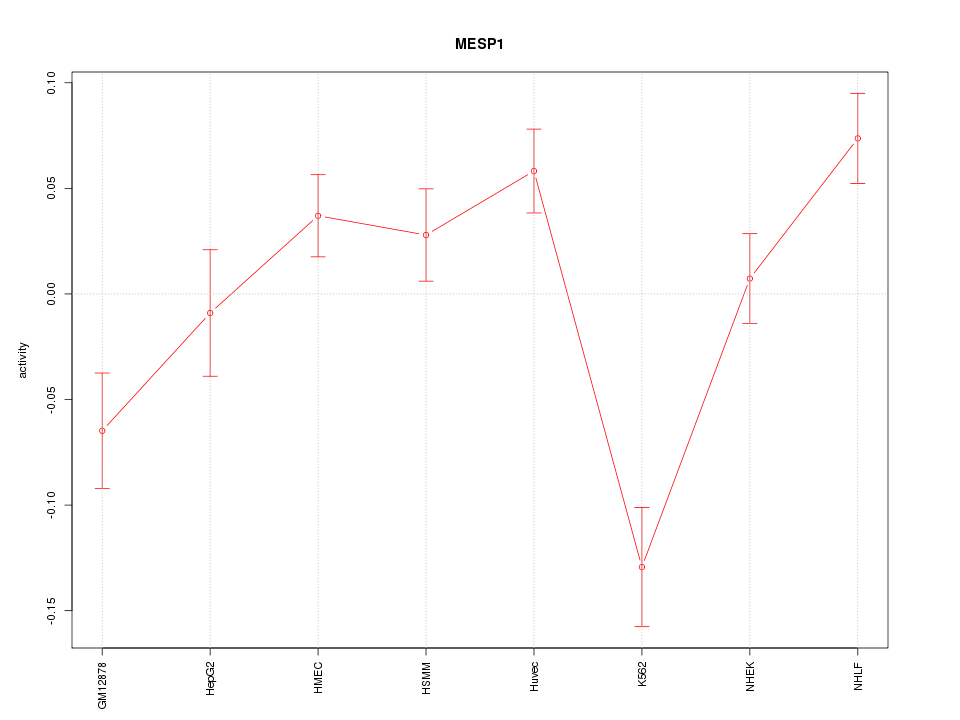
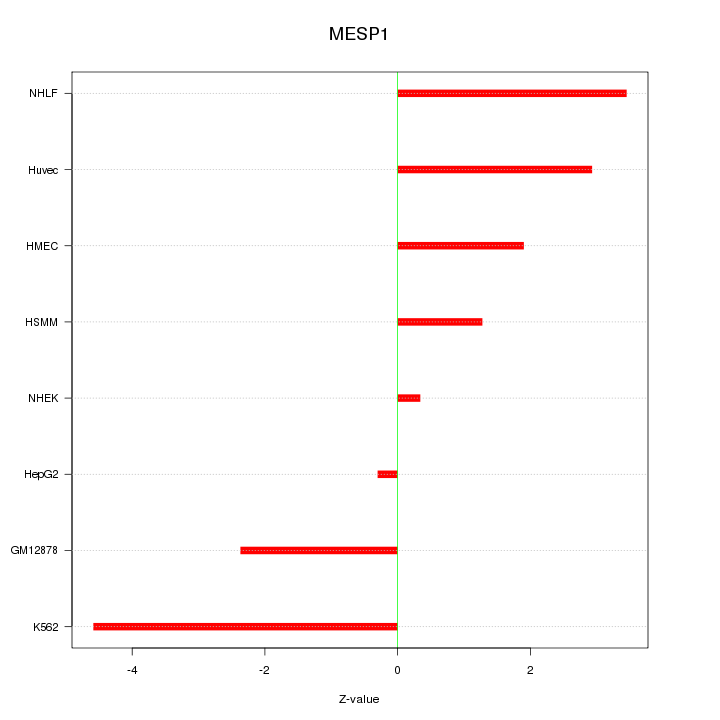
| 3.1 |
15.5 |
GO:0044854 |
membrane raft assembly(GO:0001765) nitric oxide homeostasis(GO:0033484) negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) membrane assembly(GO:0071709) |
| 1.9 |
7.8 |
GO:0030913 |
paranodal junction assembly(GO:0030913) |
| 1.2 |
5.9 |
GO:0046836 |
glycolipid transport(GO:0046836) |
| 0.9 |
2.7 |
GO:0010982 |
regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.9 |
0.9 |
GO:0003350 |
pulmonary myocardium development(GO:0003350) |
| 0.8 |
2.5 |
GO:0006051 |
mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.8 |
4.1 |
GO:0048669 |
collateral sprouting in absence of injury(GO:0048669) |
| 0.7 |
3.7 |
GO:0010993 |
ubiquitin homeostasis(GO:0010992) regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.7 |
2.1 |
GO:0033278 |
cell proliferation in midbrain(GO:0033278) |
| 0.7 |
2.8 |
GO:0046022 |
regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.5 |
2.7 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 0.5 |
1.6 |
GO:0034205 |
beta-amyloid formation(GO:0034205) |
| 0.5 |
2.0 |
GO:2001258 |
negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) negative regulation of calcium ion transmembrane transporter activity(GO:1901020) negative regulation of cation channel activity(GO:2001258) |
| 0.5 |
2.0 |
GO:0033197 |
response to vitamin E(GO:0033197) |
| 0.5 |
1.0 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.5 |
1.4 |
GO:0002541 |
activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.5 |
1.4 |
GO:0048627 |
myoblast development(GO:0048627) |
| 0.4 |
6.2 |
GO:0000052 |
citrulline metabolic process(GO:0000052) arginine catabolic process(GO:0006527) |
| 0.4 |
3.3 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.4 |
1.2 |
GO:0060689 |
cell differentiation involved in salivary gland development(GO:0060689) |
| 0.4 |
1.2 |
GO:0060054 |
positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.4 |
2.8 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.4 |
2.3 |
GO:0009649 |
entrainment of circadian clock(GO:0009649) |
| 0.4 |
1.9 |
GO:0048682 |
axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.4 |
10.3 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.3 |
3.1 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.3 |
3.8 |
GO:0098927 |
early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.3 |
1.3 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.3 |
3.6 |
GO:0010799 |
regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.3 |
1.3 |
GO:0034201 |
response to oleic acid(GO:0034201) |
| 0.3 |
0.9 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.3 |
2.1 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.3 |
0.6 |
GO:0071071 |
regulation of phosphatidylinositol biosynthetic process(GO:0010511) regulation of phospholipid biosynthetic process(GO:0071071) |
| 0.3 |
0.9 |
GO:0021592 |
fourth ventricle development(GO:0021592) |
| 0.3 |
0.9 |
GO:0019367 |
fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.3 |
4.2 |
GO:0097031 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.3 |
1.9 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.3 |
1.1 |
GO:0006663 |
platelet activating factor biosynthetic process(GO:0006663) positive regulation of vesicle fusion(GO:0031340) |
| 0.3 |
3.8 |
GO:1901797 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.3 |
2.2 |
GO:0055093 |
response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.3 |
0.8 |
GO:0002384 |
hepatic immune response(GO:0002384) |
| 0.3 |
1.9 |
GO:0010591 |
regulation of lamellipodium assembly(GO:0010591) |
| 0.2 |
4.9 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.2 |
2.6 |
GO:0007130 |
synaptonemal complex assembly(GO:0007130) |
| 0.2 |
2.6 |
GO:0010658 |
striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.2 |
0.5 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.2 |
0.5 |
GO:0060693 |
regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.2 |
0.8 |
GO:2000178 |
negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.2 |
1.9 |
GO:0071692 |
sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.2 |
1.0 |
GO:0070669 |
response to interleukin-2(GO:0070669) |
| 0.2 |
1.2 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.2 |
3.0 |
GO:0060644 |
mammary gland epithelial cell differentiation(GO:0060644) |
| 0.2 |
0.8 |
GO:0070933 |
histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.2 |
0.4 |
GO:0035106 |
operant conditioning(GO:0035106) |
| 0.2 |
1.2 |
GO:0000098 |
sulfur amino acid catabolic process(GO:0000098) |
| 0.2 |
1.3 |
GO:0072678 |
astrocyte cell migration(GO:0043615) T cell migration(GO:0072678) |
| 0.2 |
0.6 |
GO:0034770 |
histone H4-K20 methylation(GO:0034770) |
| 0.2 |
0.5 |
GO:0048706 |
embryonic skeletal system development(GO:0048706) |
| 0.2 |
2.0 |
GO:0051927 |
obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.2 |
0.5 |
GO:0045062 |
extrathymic T cell differentiation(GO:0033078) extrathymic T cell selection(GO:0045062) |
| 0.2 |
0.9 |
GO:0010544 |
negative regulation of platelet activation(GO:0010544) |
| 0.2 |
1.7 |
GO:0016540 |
protein autoprocessing(GO:0016540) |
| 0.2 |
0.8 |
GO:0019471 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 |
2.2 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 |
0.6 |
GO:0032914 |
positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 |
0.6 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 |
1.8 |
GO:0060638 |
mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.1 |
1.4 |
GO:0045329 |
amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.1 |
2.8 |
GO:0032400 |
melanosome localization(GO:0032400) |
| 0.1 |
0.1 |
GO:0022605 |
oogenesis stage(GO:0022605) |
| 0.1 |
0.4 |
GO:0006530 |
asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 |
2.8 |
GO:0050927 |
positive regulation of positive chemotaxis(GO:0050927) |
| 0.1 |
0.3 |
GO:0014887 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 |
0.7 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.1 |
1.1 |
GO:0006937 |
regulation of muscle contraction(GO:0006937) |
| 0.1 |
0.4 |
GO:0031034 |
skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament organization(GO:0031033) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 |
0.6 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 |
10.0 |
GO:0008064 |
regulation of actin polymerization or depolymerization(GO:0008064) |
| 0.1 |
1.3 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.1 |
0.5 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 |
0.6 |
GO:0001946 |
lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.1 |
1.3 |
GO:0030041 |
actin filament polymerization(GO:0030041) |
| 0.1 |
2.2 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.1 |
0.7 |
GO:0014041 |
regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.1 |
0.3 |
GO:0034086 |
maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 |
0.7 |
GO:0042693 |
muscle cell fate commitment(GO:0042693) |
| 0.1 |
0.6 |
GO:0006772 |
thiamine metabolic process(GO:0006772) |
| 0.1 |
0.6 |
GO:0070059 |
intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.1 |
0.6 |
GO:0001514 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 |
3.2 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.1 |
0.1 |
GO:0035587 |
purinergic receptor signaling pathway(GO:0035587) purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 |
7.5 |
GO:0006939 |
smooth muscle contraction(GO:0006939) |
| 0.1 |
0.7 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 |
0.5 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
0.9 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.1 |
0.6 |
GO:0003197 |
endocardial cushion development(GO:0003197) |
| 0.1 |
2.6 |
GO:0048010 |
vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.1 |
0.4 |
GO:0034287 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 |
2.5 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.1 |
0.7 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.1 |
0.5 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.1 |
0.3 |
GO:0048486 |
parasympathetic nervous system development(GO:0048486) |
| 0.1 |
1.4 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.1 |
1.1 |
GO:0002011 |
morphogenesis of an epithelial sheet(GO:0002011) |
| 0.1 |
1.8 |
GO:0042733 |
embryonic digit morphogenesis(GO:0042733) |
| 0.1 |
1.4 |
GO:0035115 |
embryonic forelimb morphogenesis(GO:0035115) |
| 0.1 |
2.0 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.1 |
0.2 |
GO:0031338 |
regulation of vesicle fusion(GO:0031338) |
| 0.1 |
0.3 |
GO:0030997 |
regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 |
0.4 |
GO:0014819 |
regulation of skeletal muscle contraction(GO:0014819) |
| 0.1 |
0.9 |
GO:0006027 |
glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 |
1.2 |
GO:0048207 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.1 |
0.4 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.1 |
0.2 |
GO:0007285 |
primary spermatocyte growth(GO:0007285) |
| 0.1 |
0.3 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.1 |
0.2 |
GO:0018874 |
benzoate metabolic process(GO:0018874) |
| 0.1 |
2.6 |
GO:0007613 |
memory(GO:0007613) |
| 0.1 |
0.3 |
GO:0042986 |
positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 |
0.3 |
GO:0044240 |
multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 |
0.9 |
GO:0006743 |
ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.1 |
0.8 |
GO:0071230 |
cellular response to amino acid stimulus(GO:0071230) |
| 0.1 |
0.1 |
GO:0035283 |
rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 |
0.3 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.1 |
0.5 |
GO:0019054 |
modulation by virus of host process(GO:0019054) modulation by symbiont of host cellular process(GO:0044068) |
| 0.1 |
1.7 |
GO:0008038 |
neuron recognition(GO:0008038) |
| 0.1 |
0.4 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.1 |
0.5 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 |
0.6 |
GO:0043537 |
negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.1 |
3.9 |
GO:0031668 |
cellular response to extracellular stimulus(GO:0031668) |
| 0.1 |
1.5 |
GO:0045740 |
positive regulation of DNA replication(GO:0045740) |
| 0.1 |
0.4 |
GO:0045542 |
positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.1 |
0.8 |
GO:0007172 |
signal complex assembly(GO:0007172) |
| 0.1 |
0.2 |
GO:0070384 |
Harderian gland development(GO:0070384) |
| 0.1 |
0.3 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.1 |
0.3 |
GO:0032203 |
telomere formation via telomerase(GO:0032203) |
| 0.1 |
0.2 |
GO:0048073 |
negative regulation of alpha-beta T cell differentiation(GO:0046639) regulation of eye pigmentation(GO:0048073) |
| 0.1 |
0.3 |
GO:0015722 |
canalicular bile acid transport(GO:0015722) |
| 0.1 |
0.1 |
GO:0042268 |
regulation of cytolysis(GO:0042268) |
| 0.1 |
0.3 |
GO:0070245 |
positive regulation of T cell apoptotic process(GO:0070234) positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 |
0.9 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 |
0.8 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 |
0.1 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.0 |
2.2 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 |
0.5 |
GO:0048488 |
synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 |
2.2 |
GO:0048704 |
embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 |
2.9 |
GO:0034332 |
adherens junction organization(GO:0034332) |
| 0.0 |
0.9 |
GO:0006942 |
regulation of striated muscle contraction(GO:0006942) |
| 0.0 |
0.2 |
GO:1902414 |
positive regulation of receptor recycling(GO:0001921) protein localization to adherens junction(GO:0071896) protein localization to cell junction(GO:1902414) |
| 0.0 |
1.8 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.3 |
GO:0045161 |
neuronal ion channel clustering(GO:0045161) |
| 0.0 |
0.4 |
GO:0006884 |
cell volume homeostasis(GO:0006884) |
| 0.0 |
0.7 |
GO:0006662 |
glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 |
0.4 |
GO:0009435 |
NAD biosynthetic process(GO:0009435) |
| 0.0 |
0.4 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 |
0.9 |
GO:0015813 |
L-glutamate transport(GO:0015813) |
| 0.0 |
1.6 |
GO:0006636 |
unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 |
1.2 |
GO:0045445 |
myoblast differentiation(GO:0045445) |
| 0.0 |
0.4 |
GO:1902591 |
vesicle coating(GO:0006901) COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) single-organism membrane budding(GO:1902591) |
| 0.0 |
2.2 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.0 |
0.2 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.0 |
1.3 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.2 |
GO:0045745 |
positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 |
2.6 |
GO:0071772 |
BMP signaling pathway(GO:0030509) response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 |
0.1 |
GO:0032074 |
negative regulation of nuclease activity(GO:0032074) |
| 0.0 |
0.5 |
GO:0070509 |
calcium ion import(GO:0070509) |
| 0.0 |
0.1 |
GO:0046341 |
CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 |
0.1 |
GO:0060046 |
regulation of acrosome reaction(GO:0060046) |
| 0.0 |
1.0 |
GO:0010332 |
response to gamma radiation(GO:0010332) |
| 0.0 |
0.4 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
0.5 |
GO:0008154 |
actin polymerization or depolymerization(GO:0008154) |
| 0.0 |
0.2 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.0 |
0.2 |
GO:0007389 |
pattern specification process(GO:0007389) |
| 0.0 |
0.5 |
GO:0000768 |
syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 |
0.2 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 |
0.1 |
GO:0001692 |
histamine metabolic process(GO:0001692) |
| 0.0 |
0.3 |
GO:0045844 |
positive regulation of striated muscle tissue development(GO:0045844) positive regulation of muscle organ development(GO:0048636) positive regulation of muscle tissue development(GO:1901863) |
| 0.0 |
1.5 |
GO:0019722 |
calcium-mediated signaling(GO:0019722) |
| 0.0 |
0.4 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.0 |
1.2 |
GO:0042059 |
negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 |
0.1 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.3 |
GO:0050434 |
positive regulation of viral transcription(GO:0050434) |
| 0.0 |
0.9 |
GO:0043473 |
pigmentation(GO:0043473) |
| 0.0 |
0.5 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.2 |
GO:0015732 |
prostaglandin transport(GO:0015732) |
| 0.0 |
0.1 |
GO:0019852 |
L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 |
0.2 |
GO:0032922 |
circadian regulation of gene expression(GO:0032922) |
| 0.0 |
0.8 |
GO:0051101 |
regulation of DNA binding(GO:0051101) |
| 0.0 |
0.3 |
GO:0043401 |
steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 |
0.7 |
GO:0040014 |
regulation of multicellular organism growth(GO:0040014) |
| 0.0 |
2.3 |
GO:0010951 |
negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 |
0.9 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 |
0.4 |
GO:0051496 |
positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 |
0.3 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
1.6 |
GO:0030521 |
androgen receptor signaling pathway(GO:0030521) |
| 0.0 |
0.1 |
GO:0051409 |
response to nitrosative stress(GO:0051409) |
| 0.0 |
0.2 |
GO:0007405 |
neuroblast proliferation(GO:0007405) |
| 0.0 |
1.0 |
GO:0006081 |
cellular aldehyde metabolic process(GO:0006081) |
| 0.0 |
0.6 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.6 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.2 |
GO:0035036 |
binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 |
0.2 |
GO:0006546 |
glycine catabolic process(GO:0006546) |
| 0.0 |
0.1 |
GO:0023041 |
neuronal signal transduction(GO:0023041) |
| 0.0 |
0.8 |
GO:0006695 |
cholesterol biosynthetic process(GO:0006695) |
| 0.0 |
0.5 |
GO:0007566 |
embryo implantation(GO:0007566) |
| 0.0 |
0.7 |
GO:0046470 |
phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 |
0.3 |
GO:0010633 |
negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 |
0.5 |
GO:0007015 |
actin filament organization(GO:0007015) |
| 0.0 |
3.3 |
GO:0001558 |
regulation of cell growth(GO:0001558) |
| 0.0 |
0.1 |
GO:0016098 |
monoterpenoid metabolic process(GO:0016098) |
| 0.0 |
0.2 |
GO:0006040 |
amino sugar metabolic process(GO:0006040) |
| 0.0 |
0.7 |
GO:0035383 |
acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 |
0.3 |
GO:0000045 |
autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 |
0.1 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.0 |
0.1 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.2 |
GO:0032727 |
positive regulation of interferon-alpha production(GO:0032727) |
| 0.0 |
0.1 |
GO:0006000 |
fructose metabolic process(GO:0006000) |
| 0.0 |
0.2 |
GO:0032784 |
regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 |
0.3 |
GO:0050680 |
negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 |
0.7 |
GO:0030049 |
muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) actin-mediated cell contraction(GO:0070252) |
| 0.0 |
0.1 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.0 |
3.4 |
GO:0051056 |
regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 |
0.1 |
GO:0002566 |
somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 |
1.8 |
GO:0050658 |
nucleic acid transport(GO:0050657) RNA transport(GO:0050658) establishment of RNA localization(GO:0051236) |
| 0.0 |
0.9 |
GO:0006090 |
pyruvate metabolic process(GO:0006090) |
| 0.0 |
0.1 |
GO:0006839 |
mitochondrial transport(GO:0006839) |
| 0.0 |
0.1 |
GO:0015780 |
nucleotide-sugar transport(GO:0015780) |