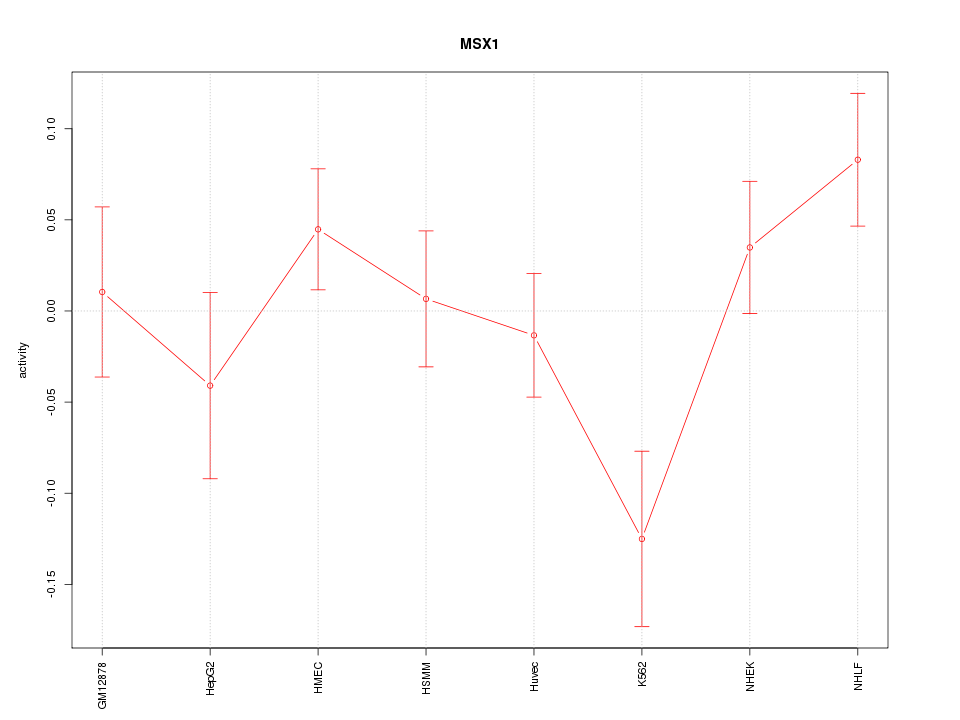
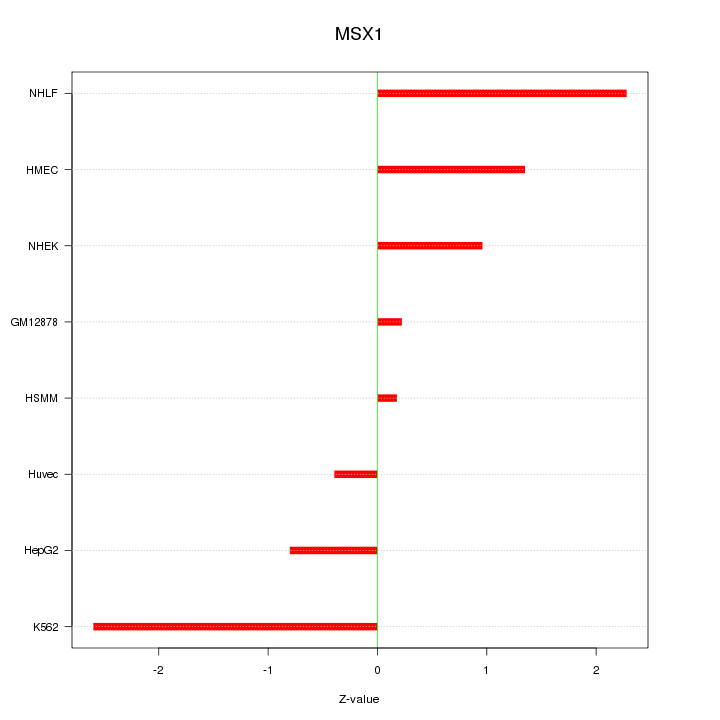
Motif ID: MSX1
Z-value: 1.395
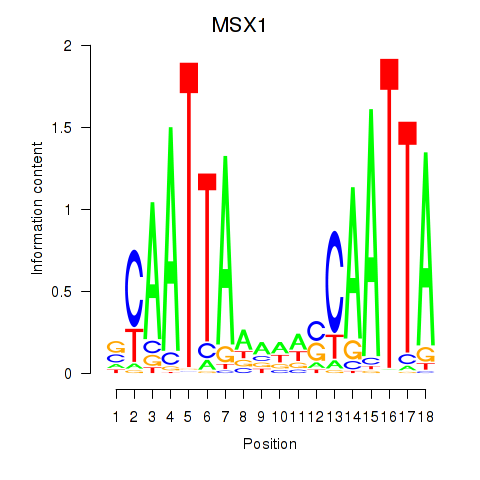
Transcription factors associated with MSX1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| MSX1 | ENSG00000163132.6 | MSX1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.4 | GO:0060414 | aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.5 | 2.0 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 2.9 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.2 | 1.3 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 3.4 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 3.6 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 14.6 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 0.4 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.1 | 1.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.5 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.7 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.5 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 1.4 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 1.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.9 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.2 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.2 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 1.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.6 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.6 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.2 | GO:0046051 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.2 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 1.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.6 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.5 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 18.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 2.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.0 | 6.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.5 | 1.4 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.4 | 1.3 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.2 | 0.5 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 2.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 3.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.0 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.5 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.5 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 8.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.6 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 1.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.4 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.3 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.0 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 3.4 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.0 | 1.3 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.6 | GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen(GO:0016705) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.4 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | SIG_CHEMOTAXIS | Genes related to chemotaxis |