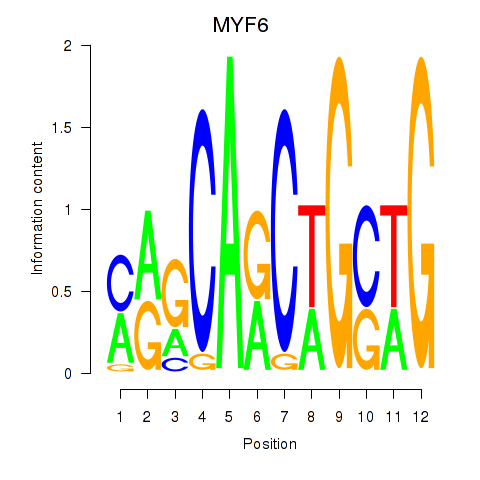
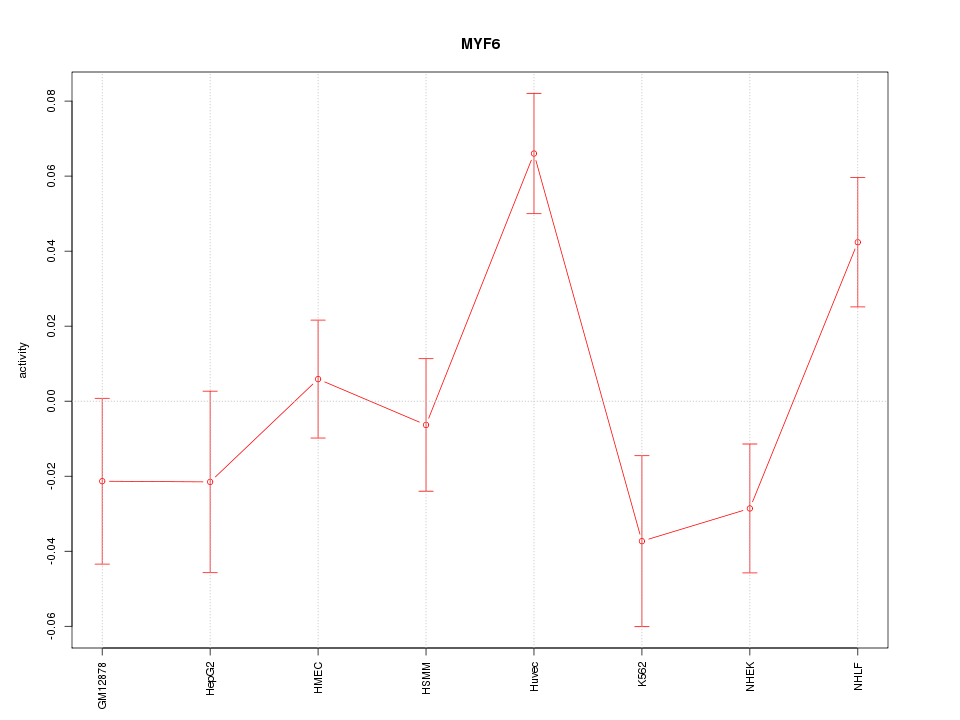
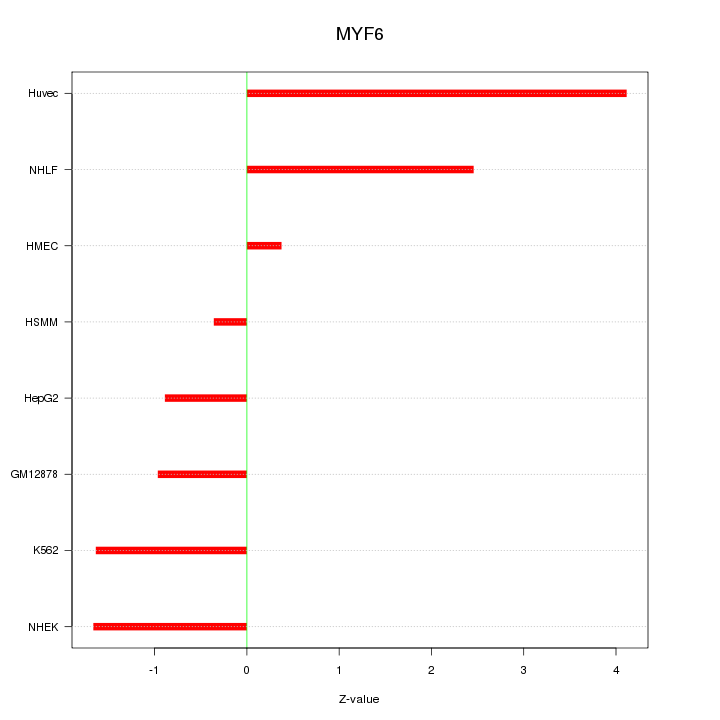
| 0.6 |
1.9 |
GO:0034036 |
purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.6 |
2.5 |
GO:0009956 |
radial pattern formation(GO:0009956) |
| 0.6 |
1.8 |
GO:0048389 |
intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) bud dilation involved in lung branching(GO:0060503) branch elongation involved in ureteric bud branching(GO:0060681) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) pulmonary artery morphogenesis(GO:0061156) cell proliferation involved in mesonephros development(GO:0061209) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) negative regulation of mesonephros development(GO:0061218) pattern specification involved in mesonephros development(GO:0061227) renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) BMP signaling pathway involved in nephric duct formation(GO:0071893) glomerular visceral epithelial cell development(GO:0072015) comma-shaped body morphogenesis(GO:0072049) regulation of branch elongation involved in ureteric bud branching(GO:0072095) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) glomerulus morphogenesis(GO:0072102) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) nephric duct formation(GO:0072179) ureter urothelium development(GO:0072190) ureter epithelial cell differentiation(GO:0072192) ureter morphogenesis(GO:0072197) metanephric comma-shaped body morphogenesis(GO:0072278) glomerular epithelial cell development(GO:0072310) negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.6 |
1.7 |
GO:0008218 |
bioluminescence(GO:0008218) |
| 0.5 |
1.4 |
GO:0060574 |
trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) intestinal epithelial cell maturation(GO:0060574) intestinal epithelial cell development(GO:0060576) |
| 0.5 |
1.4 |
GO:0042701 |
progesterone secretion(GO:0042701) |
| 0.4 |
1.6 |
GO:0010566 |
regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.4 |
2.6 |
GO:0035313 |
wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly(GO:0090504) epiboly involved in wound healing(GO:0090505) |
| 0.3 |
2.7 |
GO:0007185 |
transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.3 |
1.0 |
GO:0007389 |
pattern specification process(GO:0007389) |
| 0.3 |
1.3 |
GO:0009109 |
coenzyme catabolic process(GO:0009109) |
| 0.3 |
1.9 |
GO:1901881 |
positive regulation of protein depolymerization(GO:1901881) |
| 0.2 |
2.1 |
GO:0008354 |
germ cell migration(GO:0008354) |
| 0.2 |
1.2 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.2 |
0.8 |
GO:0009609 |
response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.2 |
0.8 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.2 |
1.0 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.2 |
1.8 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.2 |
1.2 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.2 |
1.2 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.2 |
0.8 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 |
0.5 |
GO:2000121 |
regulation of removal of superoxide radicals(GO:2000121) |
| 0.2 |
1.3 |
GO:0010799 |
regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.2 |
0.6 |
GO:0048170 |
optic nerve morphogenesis(GO:0021631) positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.2 |
0.6 |
GO:0019371 |
cyclooxygenase pathway(GO:0019371) |
| 0.2 |
0.9 |
GO:0045601 |
regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 |
0.4 |
GO:0090047 |
obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.1 |
0.6 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 |
0.4 |
GO:0060370 |
susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.1 |
1.1 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 |
0.8 |
GO:0030007 |
cellular potassium ion homeostasis(GO:0030007) negative regulation of cell volume(GO:0045794) |
| 0.1 |
1.7 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.1 |
1.4 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 |
4.3 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.1 |
0.3 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.1 |
0.4 |
GO:1903332 |
regulation of protein folding(GO:1903332) |
| 0.1 |
0.3 |
GO:0060318 |
definitive erythrocyte differentiation(GO:0060318) |
| 0.1 |
0.3 |
GO:0060316 |
positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 |
0.3 |
GO:0031133 |
regulation of axon diameter(GO:0031133) regulation of cell projection size(GO:0032536) |
| 0.1 |
3.0 |
GO:0010633 |
negative regulation of epithelial cell migration(GO:0010633) |
| 0.1 |
0.7 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |
| 0.1 |
0.8 |
GO:0022417 |
protein maturation by protein folding(GO:0022417) |
| 0.1 |
0.5 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.1 |
0.5 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 |
0.2 |
GO:0048295 |
positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.1 |
1.5 |
GO:0045749 |
obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.1 |
2.0 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.1 |
1.2 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 |
0.2 |
GO:0048549 |
positive regulation of pinocytosis(GO:0048549) |
| 0.1 |
0.8 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
1.0 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 |
0.1 |
GO:0046886 |
positive regulation of hormone metabolic process(GO:0032352) positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.1 |
0.2 |
GO:0016476 |
regulation of embryonic cell shape(GO:0016476) |
| 0.1 |
0.8 |
GO:0007130 |
synaptonemal complex assembly(GO:0007130) |
| 0.1 |
8.5 |
GO:0006367 |
transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 |
0.3 |
GO:0090343 |
positive regulation of cell aging(GO:0090343) |
| 0.1 |
1.0 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.1 |
0.2 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 |
0.7 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 |
0.9 |
GO:0046697 |
decidualization(GO:0046697) cellular response to cAMP(GO:0071320) |
| 0.1 |
1.2 |
GO:0071333 |
cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.1 |
0.7 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 |
1.5 |
GO:0042116 |
macrophage activation(GO:0042116) |
| 0.1 |
0.3 |
GO:0002903 |
negative regulation of B cell apoptotic process(GO:0002903) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.1 |
0.4 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 |
1.1 |
GO:0043567 |
regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 |
0.8 |
GO:0051927 |
obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.1 |
0.2 |
GO:0034770 |
histone H4-K20 methylation(GO:0034770) |
| 0.1 |
0.2 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.0 |
1.7 |
GO:0031532 |
actin cytoskeleton reorganization(GO:0031532) |
| 0.0 |
0.4 |
GO:0097531 |
mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 |
0.2 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 |
0.4 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
1.1 |
GO:0006662 |
glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 |
0.3 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.6 |
GO:0097031 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.7 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 |
0.2 |
GO:0021854 |
hypothalamus development(GO:0021854) |
| 0.0 |
0.6 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.0 |
0.2 |
GO:0070245 |
positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 |
0.1 |
GO:0032224 |
positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 |
0.1 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 |
0.4 |
GO:0060638 |
mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 |
0.4 |
GO:0034405 |
response to fluid shear stress(GO:0034405) |
| 0.0 |
0.2 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.1 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 |
1.7 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
0.6 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.0 |
0.5 |
GO:0006198 |
cAMP catabolic process(GO:0006198) |
| 0.0 |
0.1 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 |
0.1 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 |
0.4 |
GO:0002040 |
sprouting angiogenesis(GO:0002040) |
| 0.0 |
0.4 |
GO:0048821 |
erythrocyte development(GO:0048821) myeloid cell development(GO:0061515) |
| 0.0 |
0.1 |
GO:0046520 |
diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.0 |
0.1 |
GO:0060586 |
multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 |
0.1 |
GO:0048073 |
regulation of eye pigmentation(GO:0048073) |
| 0.0 |
0.2 |
GO:0039703 |
positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 |
0.8 |
GO:0048010 |
vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 |
0.1 |
GO:0008543 |
fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.0 |
0.5 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.2 |
GO:0000768 |
syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 |
0.7 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.1 |
GO:0019322 |
xylulose metabolic process(GO:0005997) pentose biosynthetic process(GO:0019322) |
| 0.0 |
0.0 |
GO:0007611 |
learning or memory(GO:0007611) |
| 0.0 |
0.4 |
GO:0007128 |
meiotic prophase I(GO:0007128) |
| 0.0 |
0.3 |
GO:0050718 |
positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 |
0.2 |
GO:0002011 |
morphogenesis of an epithelial sheet(GO:0002011) |
| 0.0 |
0.3 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 |
0.1 |
GO:0008212 |
mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 |
0.0 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.1 |
GO:0010658 |
negative regulation of muscle cell apoptotic process(GO:0010656) striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 |
0.1 |
GO:0035587 |
purinergic receptor signaling pathway(GO:0035587) purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 |
0.4 |
GO:0006911 |
phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 |
0.5 |
GO:0007528 |
neuromuscular junction development(GO:0007528) |
| 0.0 |
0.1 |
GO:0006707 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.1 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.7 |
GO:0015914 |
phospholipid transport(GO:0015914) |
| 0.0 |
0.3 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.0 |
0.2 |
GO:0006023 |
aminoglycan biosynthetic process(GO:0006023) |
| 0.0 |
0.2 |
GO:0090174 |
organelle membrane fusion(GO:0090174) |
| 0.0 |
0.8 |
GO:0001843 |
neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.0 |
0.3 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.0 |
GO:0019886 |
antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 |
1.8 |
GO:0032259 |
methylation(GO:0032259) |
| 0.0 |
0.1 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 |
1.2 |
GO:0007229 |
integrin-mediated signaling pathway(GO:0007229) |
| 0.0 |
0.1 |
GO:0071451 |
removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 |
0.1 |
GO:0031290 |
retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 |
0.5 |
GO:0070374 |
positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.0 |
0.1 |
GO:0098543 |
detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.0 |
0.1 |
GO:0006072 |
glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 |
0.0 |
GO:1901642 |
purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.0 |
0.8 |
GO:0016126 |
sterol biosynthetic process(GO:0016126) |
| 0.0 |
0.1 |
GO:0000394 |
RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 |
0.5 |
GO:0033572 |
ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 |
0.0 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 |
0.2 |
GO:1901661 |
quinone metabolic process(GO:1901661) |
| 0.0 |
0.1 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) NADH metabolic process(GO:0006734) |
| 0.0 |
0.3 |
GO:0007569 |
cell aging(GO:0007569) |
| 0.0 |
0.3 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.0 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.0 |
0.3 |
GO:0040014 |
regulation of multicellular organism growth(GO:0040014) |
| 0.0 |
0.1 |
GO:0045907 |
positive regulation of vasoconstriction(GO:0045907) |
| 0.0 |
0.3 |
GO:0042573 |
retinoic acid metabolic process(GO:0042573) |