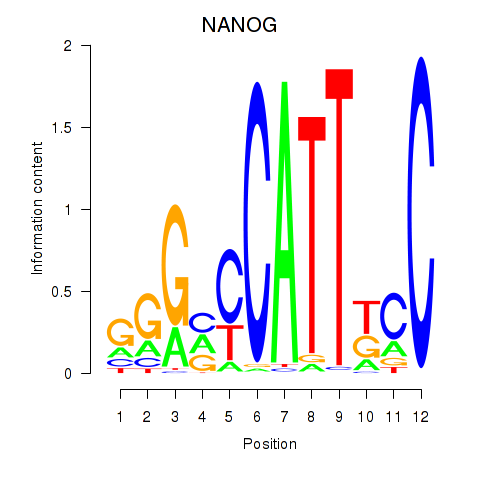
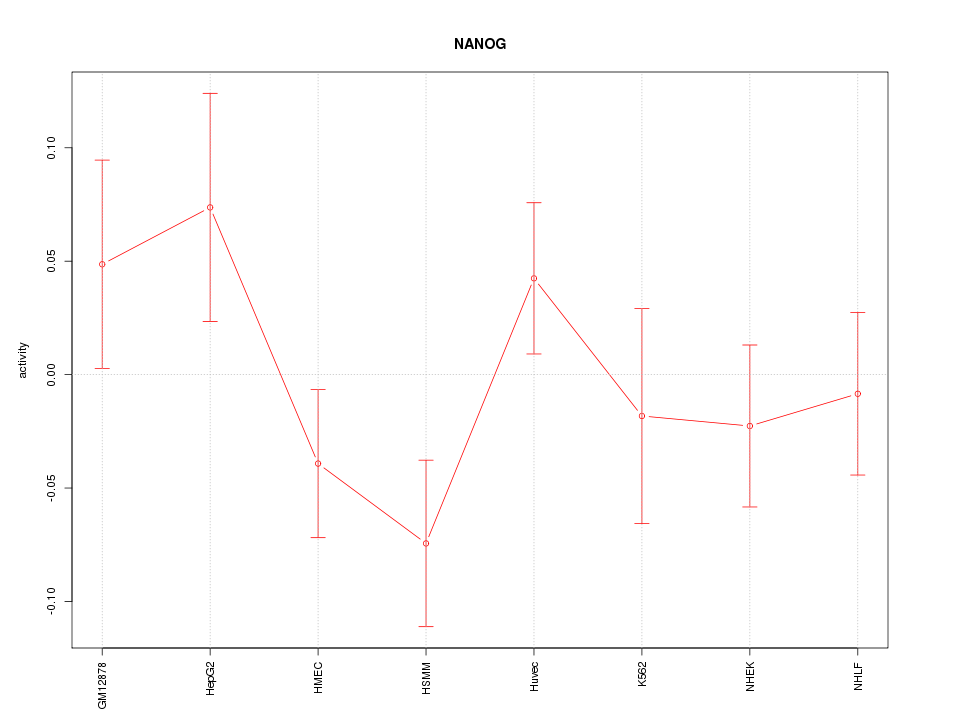
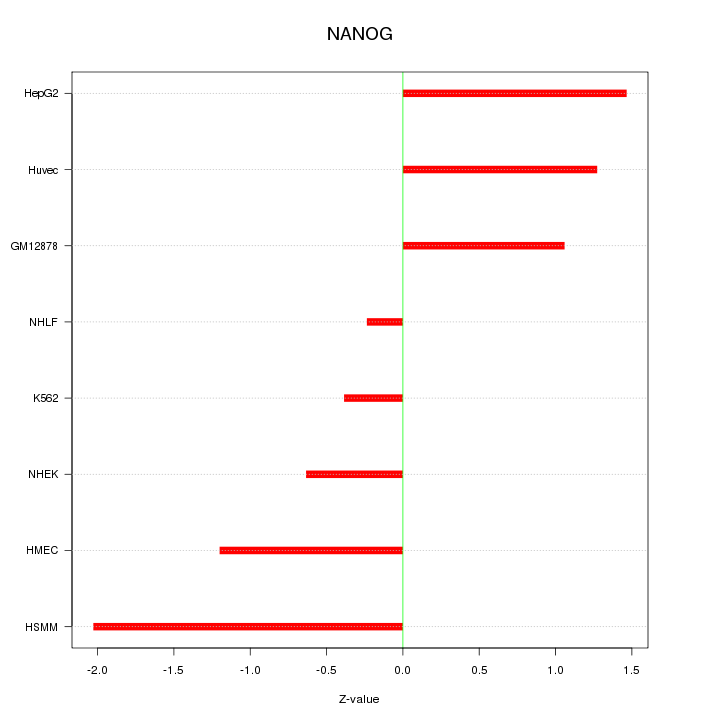
| 0.5 |
3.2 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.3 |
1.4 |
GO:0006651 |
diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 |
0.6 |
GO:0071539 |
Golgi localization(GO:0051645) protein localization to centrosome(GO:0071539) |
| 0.1 |
0.4 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.1 |
0.4 |
GO:0002678 |
positive regulation of chronic inflammatory response(GO:0002678) regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.1 |
0.7 |
GO:0010842 |
retina layer formation(GO:0010842) camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 |
0.3 |
GO:0042097 |
lymphotoxin A production(GO:0032641) interleukin-4 biosynthetic process(GO:0042097) lymphotoxin A biosynthetic process(GO:0042109) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 |
0.3 |
GO:0030037 |
actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.1 |
0.3 |
GO:0010957 |
negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 |
0.2 |
GO:0032071 |
regulation of endodeoxyribonuclease activity(GO:0032071) negative regulation of helicase activity(GO:0051097) |
| 0.1 |
1.4 |
GO:0002504 |
antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 |
0.2 |
GO:0046778 |
modification by virus of host mRNA processing(GO:0046778) |
| 0.0 |
0.1 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) response to cobalt ion(GO:0032025) |
| 0.0 |
0.2 |
GO:0032959 |
inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 |
0.2 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.0 |
0.1 |
GO:0030812 |
muscle hypertrophy in response to stress(GO:0003299) response to stimulus involved in regulation of muscle adaptation(GO:0014874) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.0 |
0.1 |
GO:0045835 |
negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 |
0.2 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 |
0.6 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 |
0.1 |
GO:0014742 |
positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 |
0.2 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.2 |
GO:0009082 |
branched-chain amino acid biosynthetic process(GO:0009082) |
| 0.0 |
0.3 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 |
1.0 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.1 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.0 |
1.2 |
GO:0001570 |
vasculogenesis(GO:0001570) |
| 0.0 |
0.2 |
GO:0001885 |
endothelial cell development(GO:0001885) |
| 0.0 |
0.7 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.0 |
0.6 |
GO:0045069 |
regulation of viral genome replication(GO:0045069) |
| 0.0 |
0.5 |
GO:0005980 |
glycogen catabolic process(GO:0005980) |
| 0.0 |
0.3 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.1 |
GO:0019321 |
pentose metabolic process(GO:0019321) |
| 0.0 |
1.5 |
GO:0006921 |
cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.0 |
0.9 |
GO:0000910 |
cytokinesis(GO:0000910) |