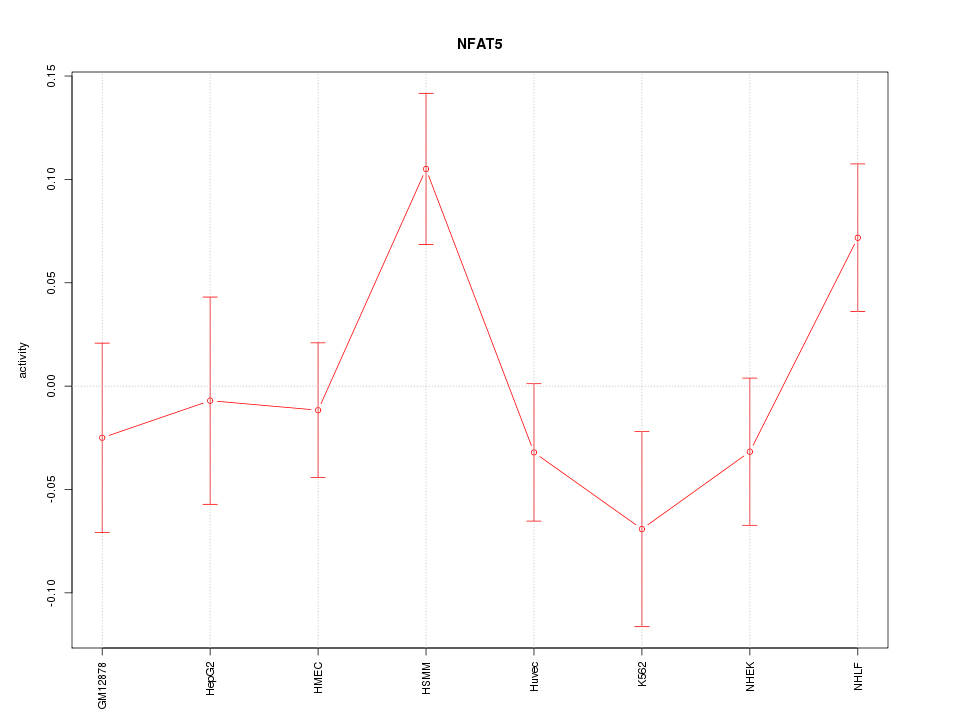
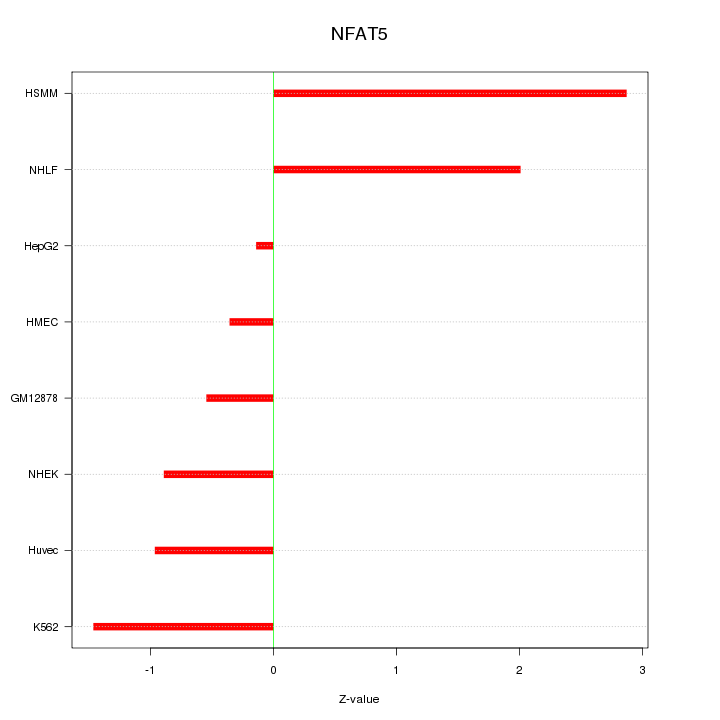
Motif ID: NFAT5
Z-value: 1.440
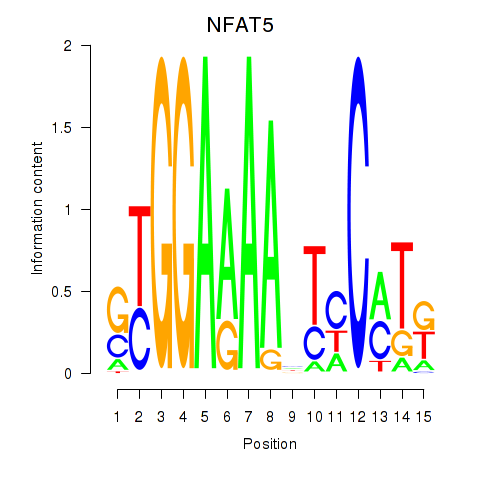
Transcription factors associated with NFAT5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NFAT5 | ENSG00000102908.16 | NFAT5 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.5 | 1.4 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.4 | 2.7 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.3 | 2.8 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.3 | 1.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.3 | 0.8 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.3 | 1.1 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.2 | 1.4 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.2 | 1.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 0.6 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.1 | 0.7 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 3.3 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 0.5 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.4 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.1 | 0.2 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.1 | 0.3 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 2.0 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 2.8 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 1.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly(GO:0090504) epiboly involved in wound healing(GO:0090505) |
| 0.0 | 2.1 | GO:0032387 | negative regulation of intracellular transport(GO:0032387) |
| 0.0 | 1.5 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.2 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 0.7 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.7 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.0 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.5 | GO:0008016 | regulation of heart contraction(GO:0008016) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 2.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 4.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 7.1 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 4.3 | GO:0031012 | extracellular matrix(GO:0031012) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.8 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.5 | 1.4 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.4 | 2.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 1.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 0.8 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 3.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 1.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 1.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 2.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 1.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.6 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 4.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |