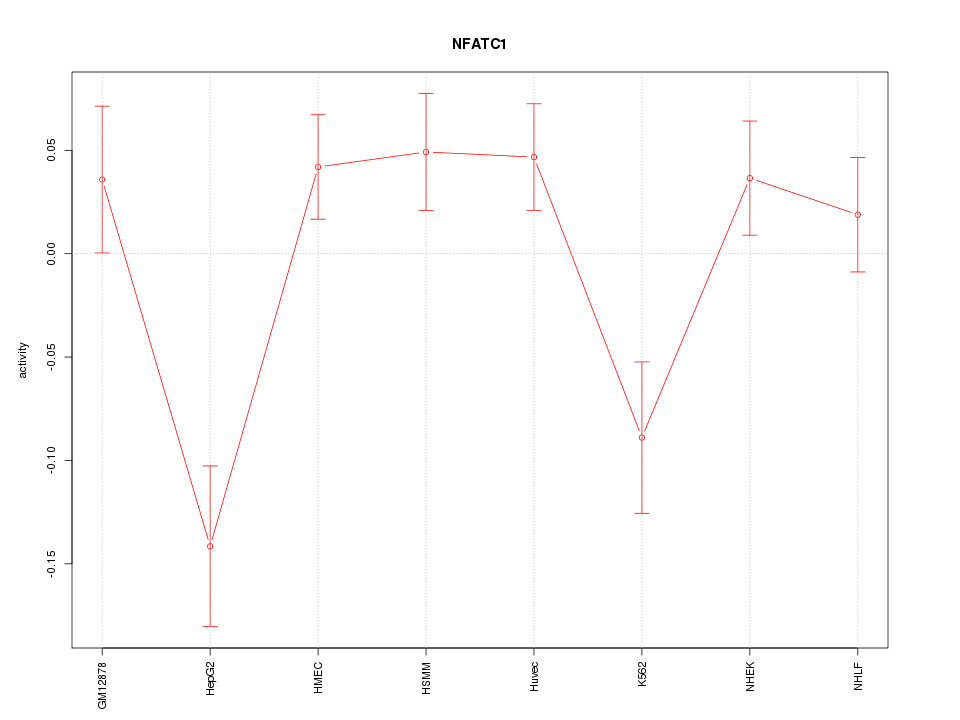
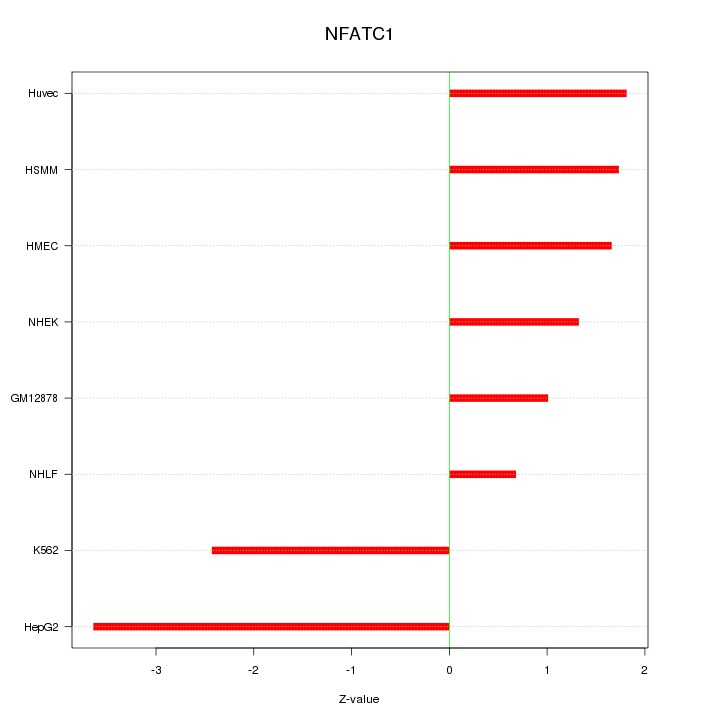
Motif ID: NFATC1
Z-value: 1.983
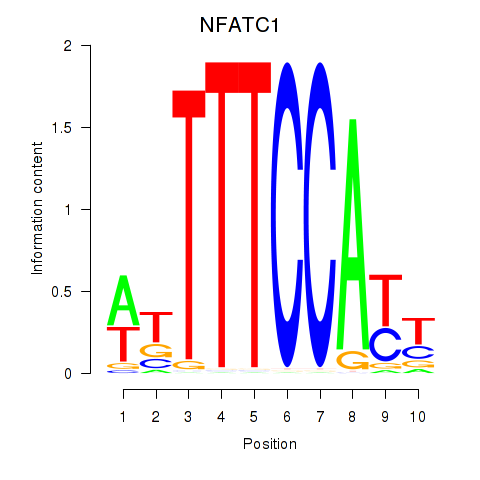
Transcription factors associated with NFATC1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NFATC1 | ENSG00000131196.13 | NFATC1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.8 | 3.1 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.7 | 4.2 | GO:2000124 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.5 | 0.5 | GO:0045908 | negative regulation of vasodilation(GO:0045908) |
| 0.5 | 1.9 | GO:0031394 | cyclooxygenase pathway(GO:0019371) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.4 | 1.8 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.4 | 2.5 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.3 | 3.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 | 0.9 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.3 | 0.9 | GO:0045726 | regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.3 | 6.0 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.3 | 2.3 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.3 | 2.3 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.2 | 1.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.2 | 1.0 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.2 | 1.6 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.2 | 0.7 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.2 | 0.9 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.2 | 2.1 | GO:0097435 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.2 | 0.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 0.9 | GO:0006789 | bilirubin conjugation(GO:0006789) biphenyl metabolic process(GO:0018879) biphenyl catabolic process(GO:0070980) |
| 0.2 | 0.5 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.2 | 0.5 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.2 | 0.6 | GO:0060649 | nipple development(GO:0060618) mammary gland bud elongation(GO:0060649) nipple morphogenesis(GO:0060658) nipple sheath formation(GO:0060659) |
| 0.1 | 0.4 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 | 0.9 | GO:0032472 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.1 | 4.2 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.1 | 0.7 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.1 | 4.6 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 2.5 | GO:0071333 | cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.1 | 0.4 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.1 | 1.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.3 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 2.4 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 1.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 1.0 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 0.4 | GO:0008298 | intracellular mRNA localization(GO:0008298) minus-end-directed organelle transport along microtubule(GO:0072385) microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 0.3 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 | 2.4 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 1.9 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 10.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 4.0 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 0.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 1.7 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.1 | 0.2 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 0.5 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.1 | 1.5 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 3.2 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.2 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.7 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.2 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.7 | GO:0044550 | secondary metabolite biosynthetic process(GO:0044550) |
| 0.0 | 0.4 | GO:0051147 | regulation of muscle cell differentiation(GO:0051147) positive regulation of muscle cell differentiation(GO:0051149) |
| 0.0 | 1.3 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 1.4 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 1.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 3.3 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.9 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.1 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0090047 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 3.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 4.5 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.2 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.6 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0071220 | response to bacterial lipoprotein(GO:0032493) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.3 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 1.4 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.0 | 0.4 | GO:1902583 | multi-organism transport(GO:0044766) transport of virus(GO:0046794) intracellular transport of virus(GO:0075733) multi-organism localization(GO:1902579) multi-organism intracellular transport(GO:1902583) |
| 0.0 | 1.1 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 4.0 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.3 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.1 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.0 | 0.7 | GO:0009408 | response to heat(GO:0009408) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 3.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.3 | 4.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 0.7 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.2 | 3.3 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.2 | 3.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 0.5 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 1.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.4 | GO:0033655 | host(GO:0018995) host cell cytoplasm(GO:0030430) host cell part(GO:0033643) host intracellular part(GO:0033646) host cell cytoplasm part(GO:0033655) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.1 | 4.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.4 | GO:0038201 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.1 | 0.4 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.4 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 4.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.4 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 2.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 3.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.7 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 1.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 2.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.4 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 1.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.9 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 7.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.8 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.4 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 4.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.8 | 10.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.5 | 1.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.5 | 1.9 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.4 | 1.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.4 | 2.3 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.3 | 5.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 4.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 1.9 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 4.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 0.6 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 1.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 2.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 7.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 0.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 0.5 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.9 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 2.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 1.5 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 3.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 6.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.5 | GO:0015216 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.1 | 0.9 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 1.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 8.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.3 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 1.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 2.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.4 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 0.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 1.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 3.5 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 7.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 1.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 4.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0004428 | obsolete inositol or phosphatidylinositol kinase activity(GO:0004428) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.3 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.6 | ST_GA13_PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.9 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.8 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.5 | ST_ADRENERGIC | Adrenergic Pathway |