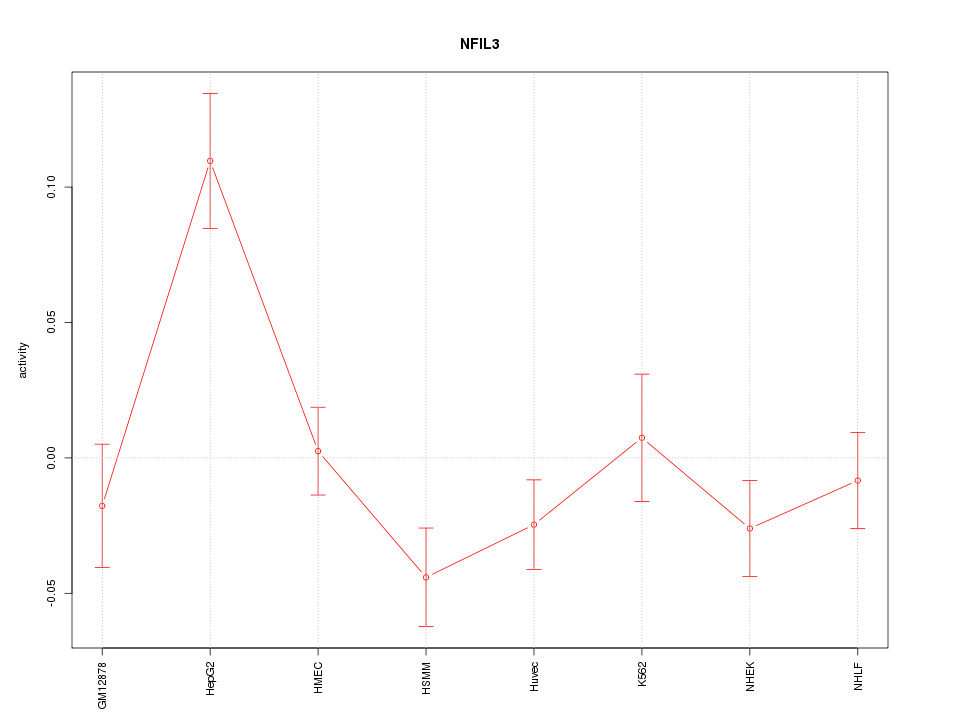
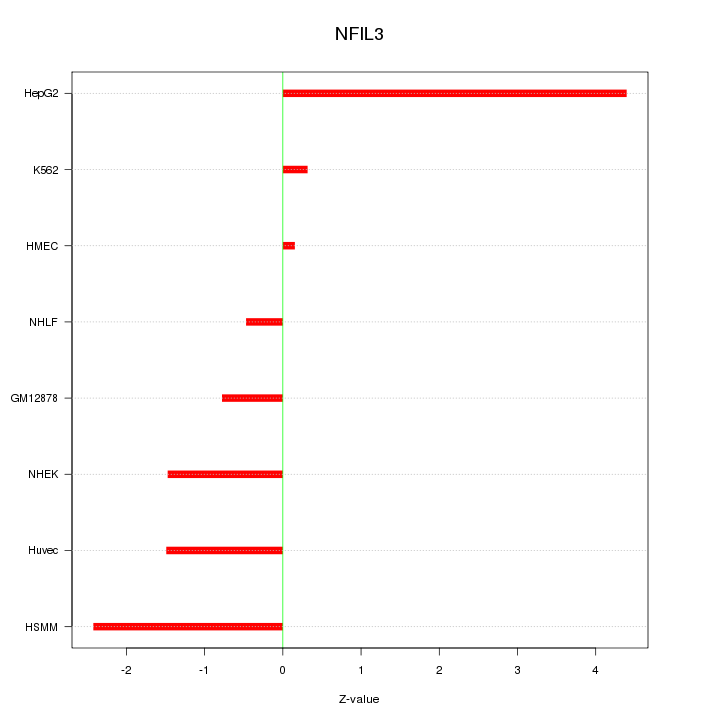
Motif ID: NFIL3
Z-value: 1.954
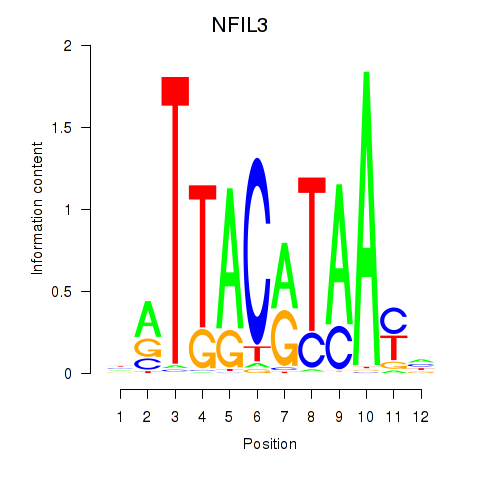
Transcription factors associated with NFIL3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NFIL3 | ENSG00000165030.3 | NFIL3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 1.3 | 4.0 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 1.1 | 3.3 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 1.1 | 3.2 | GO:0014874 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) response to stimulus involved in regulation of muscle adaptation(GO:0014874) astrocyte activation(GO:0048143) |
| 1.0 | 3.0 | GO:0090467 | response to herbicide(GO:0009635) L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.9 | 3.8 | GO:0010900 | regulation of phosphatidylcholine catabolic process(GO:0010899) negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.9 | 2.7 | GO:0061117 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.9 | 2.6 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.8 | 5.1 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.7 | 9.7 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.5 | 4.3 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.5 | 5.3 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.5 | 1.6 | GO:1903961 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.5 | 1.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.4 | 3.1 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.4 | 1.3 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.4 | 2.6 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.4 | 2.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) plasma membrane to endosome transport(GO:0048227) |
| 0.3 | 1.0 | GO:0070672 | response to interleukin-12(GO:0070671) response to interleukin-15(GO:0070672) |
| 0.3 | 1.4 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.3 | 1.0 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.3 | 1.0 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.3 | 1.2 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.3 | 0.8 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.2 | 0.7 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.2 | 0.9 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.2 | 2.1 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.2 | 2.5 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.2 | 0.2 | GO:0071223 | response to lipoteichoic acid(GO:0070391) cellular response to lipoteichoic acid(GO:0071223) |
| 0.2 | 2.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 0.6 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.2 | 0.9 | GO:0009439 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.2 | 1.9 | GO:0000050 | urea cycle(GO:0000050) |
| 0.2 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 0.7 | GO:0006408 | snRNA export from nucleus(GO:0006408) snRNA transport(GO:0051030) |
| 0.2 | 5.1 | GO:0006693 | prostaglandin metabolic process(GO:0006693) |
| 0.2 | 0.5 | GO:0010873 | positive regulation of cholesterol esterification(GO:0010873) |
| 0.2 | 0.5 | GO:0051917 | regulation of fibrinolysis(GO:0051917) |
| 0.1 | 0.6 | GO:0033622 | integrin activation(GO:0033622) regulation of integrin activation(GO:0033623) positive regulation of integrin activation(GO:0033625) |
| 0.1 | 0.7 | GO:0042519 | tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.1 | 2.0 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 4.1 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 2.3 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 | 0.4 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 0.8 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 0.3 | GO:0000114 | obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.1 | 0.3 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.1 | 2.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.9 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 0.3 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.1 | 0.4 | GO:0071801 | podosome assembly(GO:0071800) regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 2.0 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 | 0.8 | GO:0030277 | epithelial structure maintenance(GO:0010669) maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.3 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 1.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 1.0 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 | 0.7 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 0.6 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.1 | 1.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.3 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 1.1 | GO:0006560 | proline metabolic process(GO:0006560) |
| 0.1 | 0.4 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.1 | 0.3 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 0.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.1 | 0.5 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 0.6 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 2.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.2 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.1 | 0.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 4.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.5 | GO:0060325 | luteinization(GO:0001553) face morphogenesis(GO:0060325) |
| 0.1 | 1.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 1.5 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.4 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 | 0.6 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.1 | 0.7 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.1 | 3.6 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.1 | 1.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.2 | GO:0035269 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.1 | 0.9 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.8 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.2 | GO:0042511 | positive regulation of tyrosine phosphorylation of Stat1 protein(GO:0042511) |
| 0.1 | 1.2 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 1.8 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.3 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.2 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.1 | 0.7 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 0.5 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.6 | GO:0070302 | regulation of stress-activated MAPK cascade(GO:0032872) regulation of stress-activated protein kinase signaling cascade(GO:0070302) |
| 0.1 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.6 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.4 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.3 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.3 | GO:0030147 | obsolete natriuresis(GO:0030147) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.4 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.1 | GO:0001845 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.1 | GO:0001832 | blastocyst growth(GO:0001832) inner cell mass cell proliferation(GO:0001833) positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 0.3 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.0 | 0.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.5 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.9 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 1.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.3 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.4 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.2 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:0010882 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 1.1 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 1.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 2.7 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 2.3 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.2 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.3 | GO:0009750 | response to fructose(GO:0009750) |
| 0.0 | 0.3 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.2 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.4 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.5 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.2 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.2 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:0045822 | negative regulation of heart contraction(GO:0045822) |
| 0.0 | 0.3 | GO:0030163 | protein catabolic process(GO:0030163) |
| 0.0 | 0.4 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) iris morphogenesis(GO:0061072) cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.0 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.0 | 0.1 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.0 | 0.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.9 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.2 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.1 | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity(GO:0002888) positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.3 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.5 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.0 | 0.1 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.3 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.6 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 2.5 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.6 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.0 | 0.4 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.3 | GO:0009451 | RNA modification(GO:0009451) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.3 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 | 0.2 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.1 | GO:0010430 | ethanol catabolic process(GO:0006068) fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.4 | GO:0050709 | negative regulation of protein secretion(GO:0050709) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.3 | GO:0060444 | branching involved in mammary gland duct morphogenesis(GO:0060444) |
| 0.0 | 0.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.7 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.3 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.0 | 0.1 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.3 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.2 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:0043303 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) diol metabolic process(GO:0034311) |
| 0.0 | 3.1 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.0 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.2 | GO:0010332 | response to gamma radiation(GO:0010332) |
| 0.0 | 0.4 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.0 | 0.2 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.5 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.0 | GO:0071474 | cellular water homeostasis(GO:0009992) cellular hyperosmotic response(GO:0071474) |
| 0.0 | 0.1 | GO:0002860 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 10.7 | GO:0006629 | lipid metabolic process(GO:0006629) |
| 0.0 | 0.1 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.5 | GO:0007272 | ensheathment of neurons(GO:0007272) axon ensheathment(GO:0008366) |
| 0.0 | 0.1 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.0 | 0.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.5 | 5.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.5 | 2.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.4 | 1.9 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.4 | 6.4 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.4 | 5.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 3.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 0.8 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 1.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 0.8 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.2 | 1.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 0.5 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.2 | 0.6 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.1 | 1.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.4 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 2.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 1.0 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 23.5 | GO:0005792 | obsolete microsome(GO:0005792) |
| 0.1 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 1.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 1.0 | GO:0019861 | obsolete flagellum(GO:0019861) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 4.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.7 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0001940 | female pronucleus(GO:0001939) male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.2 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 2.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 11.0 | GO:0005625 | obsolete soluble fraction(GO:0005625) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.1 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.5 | GO:0012506 | vesicle membrane(GO:0012506) cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.2 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 5.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 5.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0044420 | extracellular matrix component(GO:0044420) |
| 0.0 | 0.2 | GO:0005858 | axonemal dynein complex(GO:0005858) axoneme part(GO:0044447) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 1.4 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.2 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.5 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) cytoplasmic side of membrane(GO:0098562) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.5 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 10.0 | GO:0047042 | androsterone dehydrogenase (B-specific) activity(GO:0047042) |
| 1.8 | 7.2 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.9 | 3.7 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.8 | 2.5 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.8 | 6.3 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.8 | 3.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.7 | 25.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.5 | 1.9 | GO:0019862 | IgA binding(GO:0019862) |
| 0.4 | 2.9 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.4 | 5.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 1.9 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 1.9 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.4 | 2.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.4 | 4.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.4 | 2.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.3 | 5.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.3 | 2.0 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.3 | 1.0 | GO:0070283 | lipoate-protein ligase activity(GO:0016979) lipoate synthase activity(GO:0016992) radical SAM enzyme activity(GO:0070283) |
| 0.3 | 4.0 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.2 | 0.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 0.9 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.2 | 2.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 0.7 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.2 | 2.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 0.8 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.2 | 3.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 1.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 0.8 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.2 | 0.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.2 | 0.9 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 1.5 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 1.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.2 | 3.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 3.0 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.2 | 4.8 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.2 | 0.6 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.1 | 0.4 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 1.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 2.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.8 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 0.4 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.1 | 0.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.5 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 1.3 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 0.5 | GO:0008907 | integrase activity(GO:0008907) |
| 0.1 | 1.0 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.3 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.4 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.3 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 0.6 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.7 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 1.5 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 0.7 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.1 | 4.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.3 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.7 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.1 | 0.8 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.9 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 0.3 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 1.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.7 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 0.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 1.0 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 3.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) fatty acid ligase activity(GO:0015645) |
| 0.1 | 1.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.2 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.1 | 0.2 | GO:0050833 | pyruvate secondary active transmembrane transporter activity(GO:0005477) pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.2 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.4 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 0.4 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 0.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.2 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 3.4 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.7 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.3 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0043426 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.2 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.8 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 5.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.5 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 2.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 1.7 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.2 | GO:0060590 | ATPase regulator activity(GO:0060590) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.0 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 2.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.2 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0030284 | nitric-oxide synthase regulator activity(GO:0030235) estrogen receptor activity(GO:0030284) estrogen response element binding(GO:0034056) |
| 0.0 | 0.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.6 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.2 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | ST_STAT3_PATHWAY | STAT3 Pathway |
| 0.0 | 0.7 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.0 | 0.7 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.8 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | ST_B_CELL_ANTIGEN_RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.2 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.7 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.3 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |