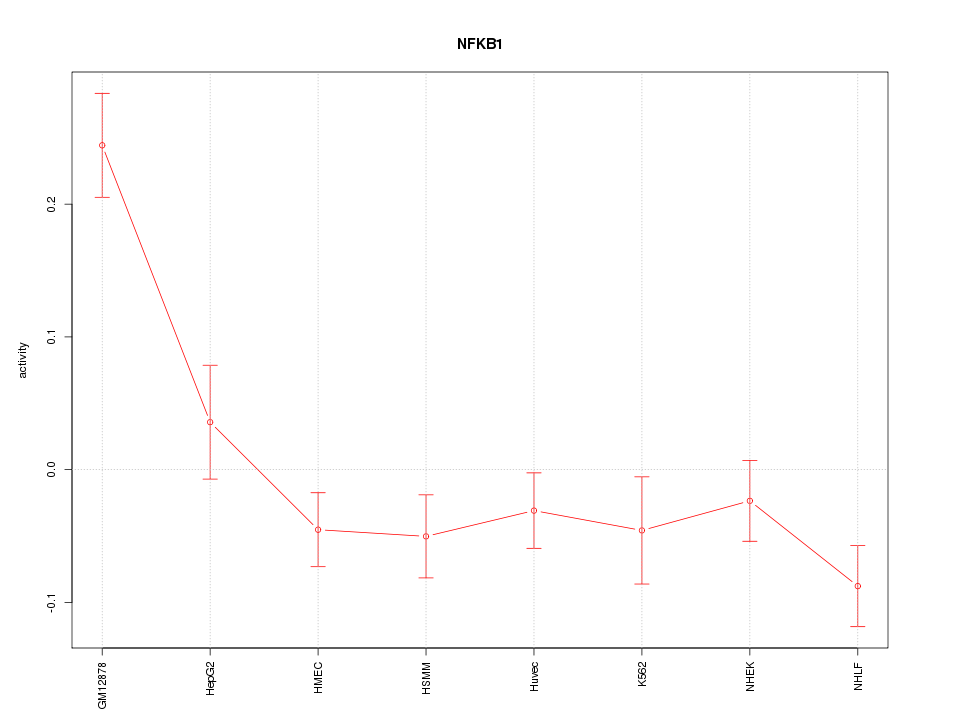
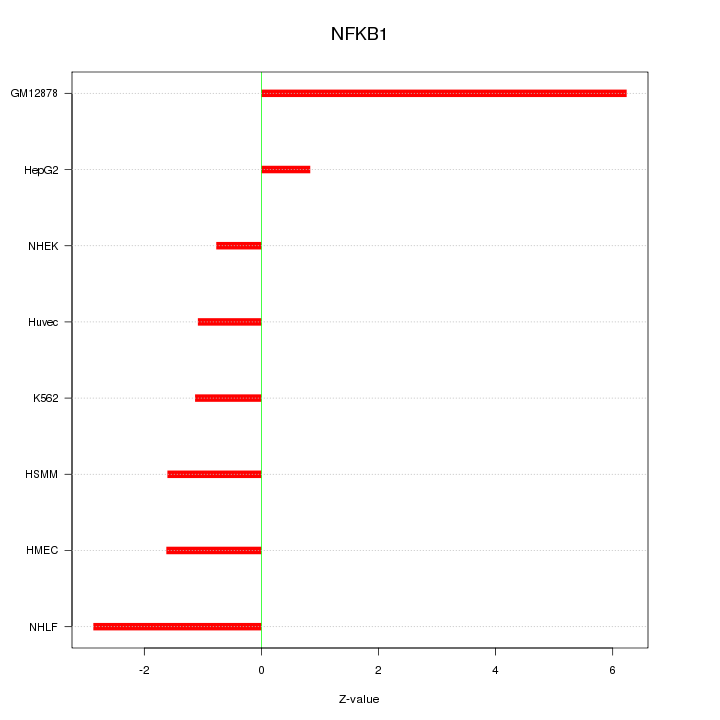
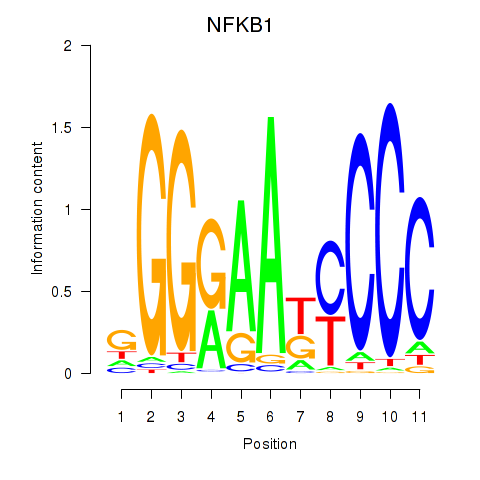
Motif ID: NFKB1
Z-value: 2.649
Transcription factors associated with NFKB1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NFKB1 | ENSG00000109320.7 | NFKB1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 2.7 | 8.0 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 1.4 | 5.6 | GO:0070842 | aggresome assembly(GO:0070842) |
| 1.3 | 5.3 | GO:1904037 | positive regulation of epithelial cell apoptotic process(GO:1904037) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 1.3 | 3.9 | GO:0070427 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 1.1 | 3.4 | GO:0002678 | positive regulation of chronic inflammatory response(GO:0002678) regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 1.1 | 3.2 | GO:0045062 | extrathymic T cell differentiation(GO:0033078) extrathymic T cell selection(GO:0045062) |
| 1.1 | 6.3 | GO:0060263 | regulation of hydrogen peroxide metabolic process(GO:0010310) regulation of respiratory burst(GO:0060263) |
| 1.0 | 7.8 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.8 | 1.5 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.8 | 6.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.7 | 8.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.7 | 4.4 | GO:0032916 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.6 | 0.6 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.6 | 5.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.5 | 3.7 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.5 | 2.6 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.5 | 11.4 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.5 | 2.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.4 | 2.2 | GO:0032329 | serine transport(GO:0032329) |
| 0.4 | 6.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.4 | 1.6 | GO:0002268 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.4 | 7.9 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.3 | 1.0 | GO:0060024 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) rhythmic synaptic transmission(GO:0060024) |
| 0.3 | 9.5 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.3 | 0.5 | GO:0032825 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.2 | 1.6 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.2 | 1.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 2.8 | GO:0032613 | interleukin-10 production(GO:0032613) |
| 0.2 | 1.0 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.2 | 1.7 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.2 | 1.2 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.2 | 0.8 | GO:0032674 | interleukin-5 production(GO:0032634) regulation of interleukin-5 production(GO:0032674) positive regulation of interleukin-5 production(GO:0032754) |
| 0.2 | 1.9 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.1 | 11.3 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.1 | 0.6 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.1 | 0.4 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 0.6 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 2.7 | GO:0051925 | obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.1 | 0.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.5 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.7 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.5 | GO:0070245 | positive regulation of lymphocyte apoptotic process(GO:0070230) positive regulation of T cell apoptotic process(GO:0070234) positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 0.3 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 1.2 | GO:0006984 | ER-nucleus signaling pathway(GO:0006984) |
| 0.1 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 1.0 | GO:0032608 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) |
| 0.1 | 1.2 | GO:0090003 | regulation of establishment of protein localization to plasma membrane(GO:0090003) regulation of protein localization to plasma membrane(GO:1903076) regulation of protein localization to cell periphery(GO:1904375) |
| 0.1 | 0.4 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.6 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 4.1 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 1.3 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 3.4 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 4.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 2.0 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 0.1 | GO:1901142 | insulin processing(GO:0030070) insulin metabolic process(GO:1901142) |
| 0.0 | 10.1 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 2.0 | GO:0001892 | embryonic placenta development(GO:0001892) |
| 0.0 | 0.3 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 1.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0042789 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 2.5 | GO:0061025 | membrane fusion(GO:0061025) |
| 0.0 | 1.1 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.4 | GO:0040023 | establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.9 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 1.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.7 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 4.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.2 | GO:0009629 | response to gravity(GO:0009629) otolith development(GO:0048840) |
| 0.0 | 0.0 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.0 | 0.9 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.1 | GO:0051000 | positive regulation of nitric-oxide synthase activity(GO:0051000) |
| 0.0 | 0.3 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.2 | GO:0015669 | gas transport(GO:0015669) |
| 0.0 | 2.2 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.4 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.1 | GO:0009820 | alkaloid metabolic process(GO:0009820) monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.5 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.0 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 4.8 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 9.5 | GO:0019815 | immunoglobulin complex(GO:0019814) B cell receptor complex(GO:0019815) |
| 1.0 | 8.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 1.0 | 21.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.7 | 4.2 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.5 | 6.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.3 | 3.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 5.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.3 | 2.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.3 | 7.4 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.3 | 1.6 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 6.9 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.1 | 2.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 6.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 3.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 2.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.6 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 1.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 3.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 9.2 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 4.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 6.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.7 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 3.1 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) cytoplasmic side of membrane(GO:0098562) |
| 0.0 | 2.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.7 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 3.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.8 | 11.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.7 | 8.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.5 | 8.0 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.5 | 3.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.4 | 12.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.4 | 3.7 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.4 | 2.1 | GO:0046980 | oligopeptide-transporting ATPase activity(GO:0015421) tapasin binding(GO:0046980) |
| 0.4 | 5.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.4 | 1.5 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
| 0.3 | 5.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 2.3 | GO:0042834 | peptidoglycan binding(GO:0042834) CARD domain binding(GO:0050700) |
| 0.3 | 6.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 1.2 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.2 | 3.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 5.7 | GO:0043621 | protein self-association(GO:0043621) |
| 0.2 | 1.6 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.2 | 5.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 4.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 3.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.6 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.3 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 1.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.7 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.5 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 2.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 12.9 | GO:0003702 | obsolete RNA polymerase II transcription factor activity(GO:0003702) |
| 0.1 | 0.2 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 2.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 2.5 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 4.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 12.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 4.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 9.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 1.2 | GO:0016566 | obsolete specific transcriptional repressor activity(GO:0016566) |
| 0.0 | 1.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 2.8 | GO:0010843 | obsolete promoter binding(GO:0010843) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.5 | GO:0016564 | obsolete transcription repressor activity(GO:0016564) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 4.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.0 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 11.0 | GO:0003700 | transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 25.8 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.2 | 11.0 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 2.3 | ST_TYPE_I_INTERFERON_PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.5 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.3 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |