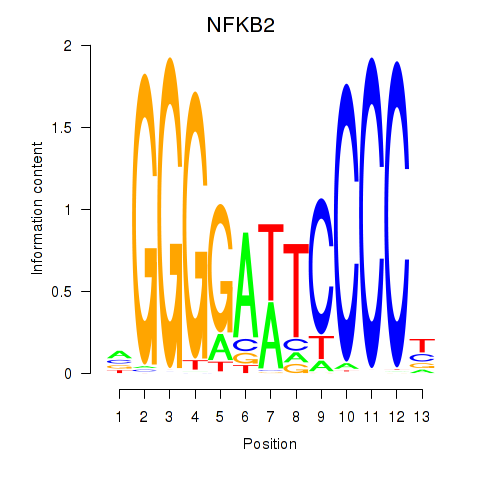
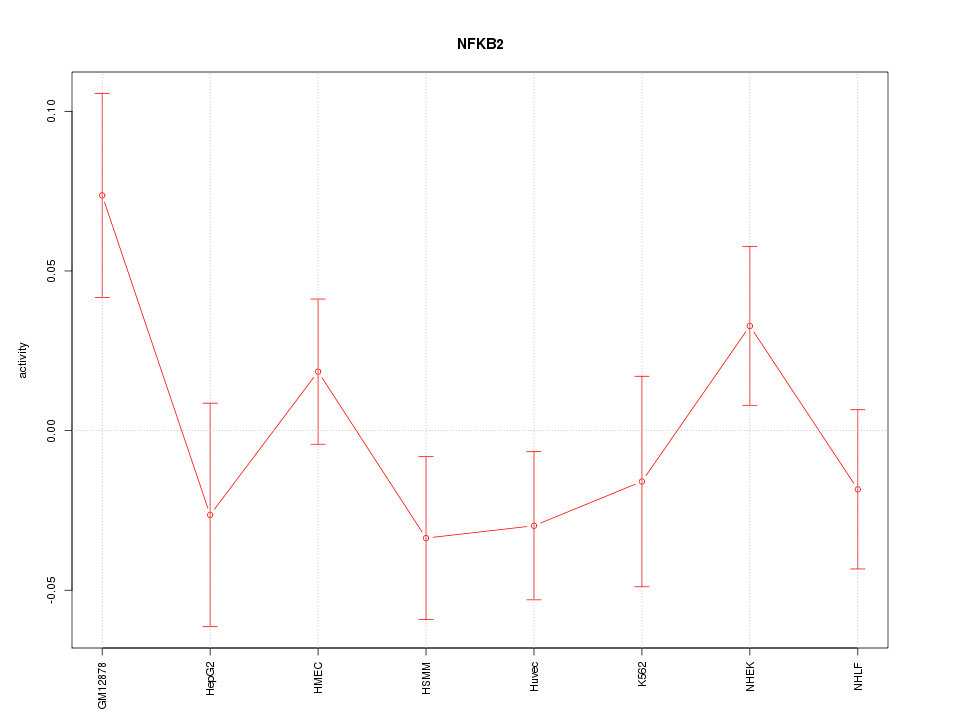
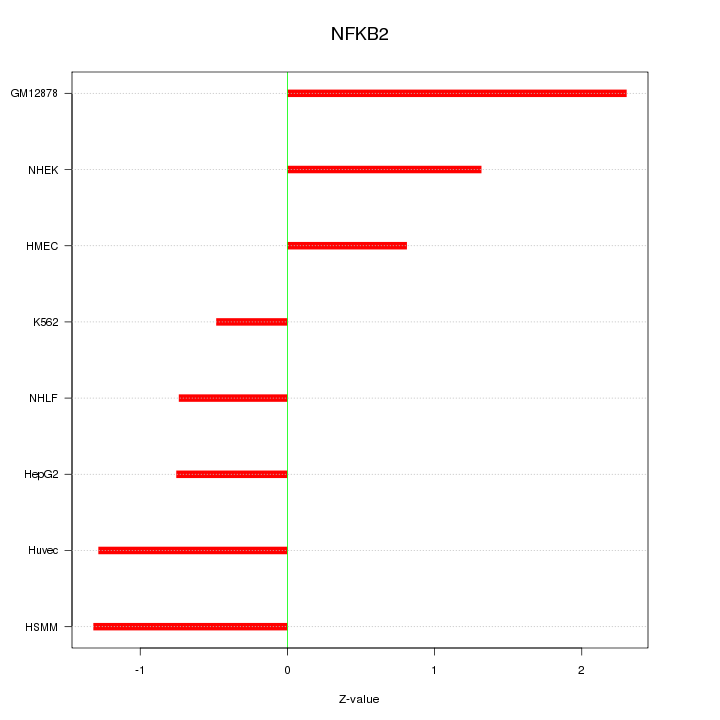
| 0.6 |
3.7 |
GO:0060263 |
regulation of respiratory burst(GO:0060263) |
| 0.5 |
1.6 |
GO:0071460 |
negative regulation of necrotic cell death(GO:0060547) cellular response to cell-matrix adhesion(GO:0071460) |
| 0.4 |
2.7 |
GO:0034616 |
response to laminar fluid shear stress(GO:0034616) |
| 0.4 |
1.9 |
GO:0032959 |
inositol trisphosphate biosynthetic process(GO:0032959) positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of cation channel activity(GO:2001259) |
| 0.4 |
1.1 |
GO:0072677 |
eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.3 |
3.7 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.3 |
1.0 |
GO:0071603 |
negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) endothelial cell-cell adhesion(GO:0071603) |
| 0.3 |
0.9 |
GO:0002678 |
positive regulation of chronic inflammatory response(GO:0002678) regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.3 |
0.8 |
GO:0009635 |
response to herbicide(GO:0009635) |
| 0.2 |
0.9 |
GO:0032849 |
positive regulation of cellular pH reduction(GO:0032849) |
| 0.2 |
1.8 |
GO:0032713 |
negative regulation of interleukin-4 production(GO:0032713) |
| 0.2 |
4.6 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.2 |
3.7 |
GO:0002504 |
antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.2 |
0.8 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.2 |
1.4 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 |
0.4 |
GO:0009134 |
nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 |
0.1 |
GO:0003307 |
regulation of Wnt signaling pathway involved in heart development(GO:0003307) |
| 0.1 |
0.5 |
GO:0060292 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) negative regulation of phagocytosis(GO:0050765) long term synaptic depression(GO:0060292) |
| 0.1 |
0.3 |
GO:0033197 |
response to vitamin E(GO:0033197) |
| 0.1 |
1.6 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.1 |
0.6 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) |
| 0.1 |
2.1 |
GO:0050732 |
negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.1 |
2.8 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.1 |
0.5 |
GO:0022614 |
membrane to membrane docking(GO:0022614) |
| 0.1 |
0.2 |
GO:0070427 |
positive regulation of immature T cell proliferation(GO:0033091) nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) response to interleukin-12(GO:0070671) |
| 0.1 |
0.6 |
GO:0051294 |
establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.1 |
0.9 |
GO:0015939 |
pantothenate metabolic process(GO:0015939) |
| 0.1 |
0.2 |
GO:0034629 |
cellular protein complex localization(GO:0034629) |
| 0.1 |
0.6 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.1 |
0.2 |
GO:0002268 |
follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.0 |
0.3 |
GO:0006295 |
nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 |
0.4 |
GO:0043545 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.5 |
GO:0045741 |
positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 |
0.3 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.0 |
0.3 |
GO:0006477 |
protein sulfation(GO:0006477) |
| 0.0 |
0.3 |
GO:0045063 |
T-helper 1 cell differentiation(GO:0045063) |
| 0.0 |
0.4 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.0 |
0.4 |
GO:0070189 |
kynurenine metabolic process(GO:0070189) |
| 0.0 |
0.6 |
GO:0060644 |
mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 |
0.9 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 |
0.2 |
GO:0034201 |
response to oleic acid(GO:0034201) |
| 0.0 |
0.2 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 |
0.2 |
GO:0030186 |
melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 |
0.2 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.0 |
0.2 |
GO:0045350 |
interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 |
0.1 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 |
0.2 |
GO:1901503 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 |
0.2 |
GO:0048296 |
isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 |
0.1 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.0 |
1.0 |
GO:0050830 |
defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 |
0.7 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.1 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.0 |
0.1 |
GO:0097237 |
autophagic cell death(GO:0048102) cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 |
0.0 |
GO:1903514 |
regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.0 |
0.7 |
GO:0007620 |
copulation(GO:0007620) |
| 0.0 |
2.7 |
GO:0008277 |
regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 |
0.2 |
GO:0043497 |
regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 |
1.2 |
GO:0032480 |
negative regulation of type I interferon production(GO:0032480) |
| 0.0 |
0.4 |
GO:0007029 |
endoplasmic reticulum organization(GO:0007029) |
| 0.0 |
0.1 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 |
0.1 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.0 |
0.5 |
GO:0030838 |
positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 |
0.1 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 |
0.8 |
GO:0014070 |
response to organic cyclic compound(GO:0014070) |
| 0.0 |
0.6 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.0 |
0.4 |
GO:0033209 |
tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 |
0.1 |
GO:0016559 |
peroxisome membrane biogenesis(GO:0016557) peroxisome fission(GO:0016559) |
| 0.0 |
0.5 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.1 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 |
0.2 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 |
0.2 |
GO:0046902 |
regulation of mitochondrial membrane permeability(GO:0046902) regulation of membrane permeability(GO:0090559) |
| 0.0 |
1.0 |
GO:0051092 |
positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 |
0.1 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 |
0.1 |
GO:0060351 |
cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 |
1.5 |
GO:0000216 |
obsolete M/G1 transition of mitotic cell cycle(GO:0000216) |
| 0.0 |
0.2 |
GO:0040023 |
establishment of nucleus localization(GO:0040023) |
| 0.0 |
0.1 |
GO:2000171 |
negative regulation of dendrite development(GO:2000171) |
| 0.0 |
0.3 |
GO:0016254 |
preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 |
0.1 |
GO:0032528 |
microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 |
0.6 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.0 |
0.4 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.0 |
0.1 |
GO:0032916 |
transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 |
0.2 |
GO:1903078 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 |
0.1 |
GO:0006491 |
N-glycan processing(GO:0006491) |