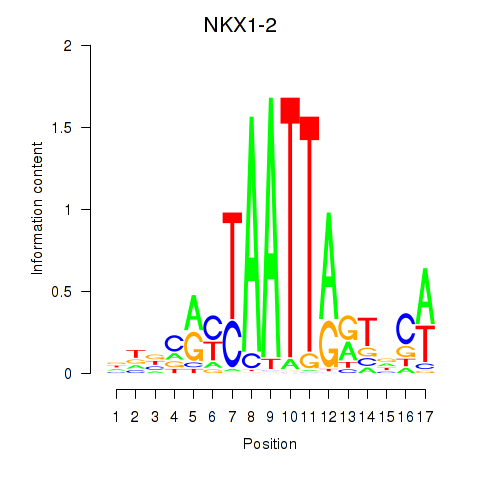
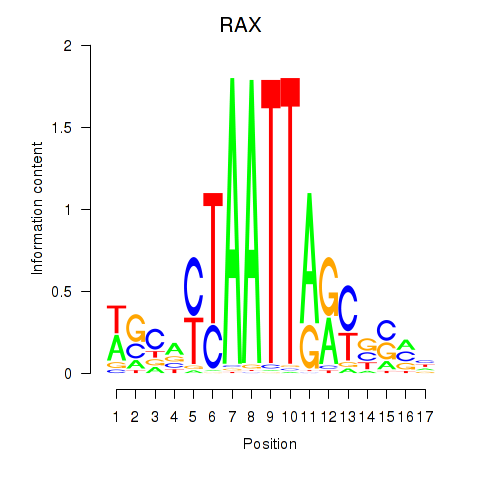
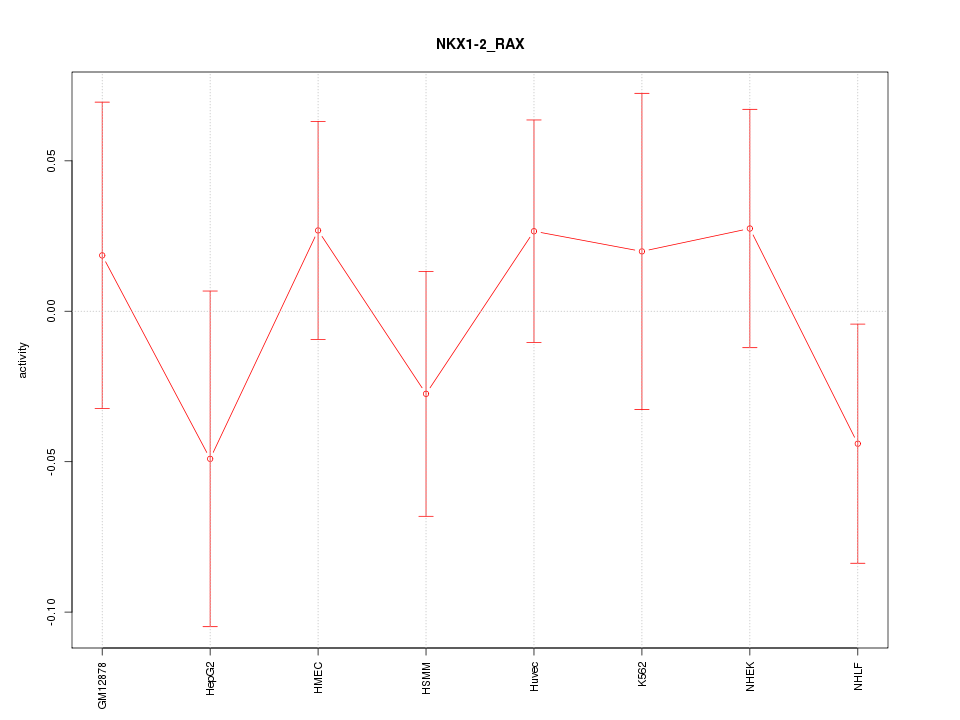
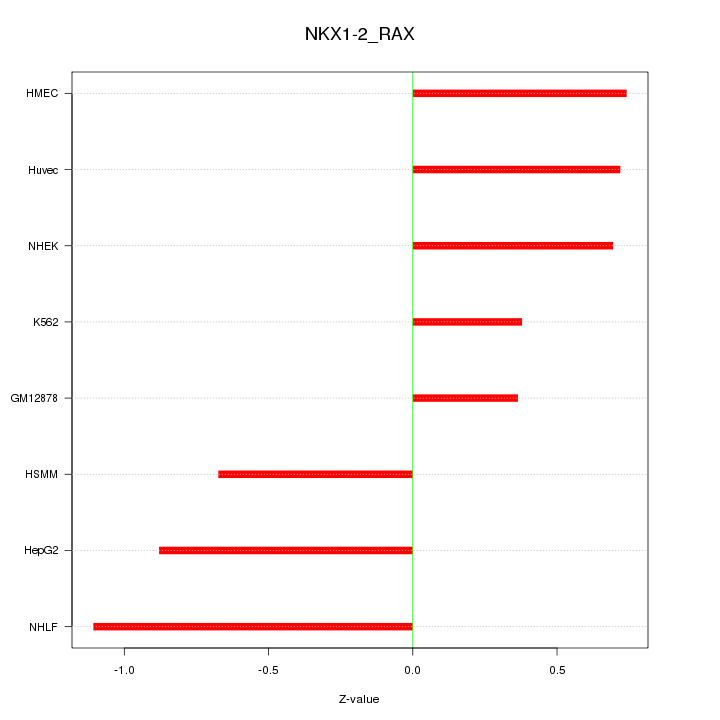
|
chr11_-_62521614
|
1.871
|
ENST00000527994.1
ENST00000394807.3
|
ZBTB3
|
zinc finger and BTB domain containing 3
|
|
chr7_-_87856280
|
1.562
|
ENST00000490437.1
ENST00000431660.1
|
SRI
|
sorcin
|
|
chr7_-_87856303
|
1.546
|
ENST00000394641.3
|
SRI
|
sorcin
|
|
chr4_-_25865159
|
1.440
|
ENST00000502949.1
ENST00000264868.5
ENST00000513691.1
ENST00000514872.1
|
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans)
|
|
chr3_-_151034734
|
1.407
|
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87
|
|
chr5_+_66300446
|
0.751
|
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr6_+_34204642
|
0.584
|
ENST00000347617.6
ENST00000401473.3
ENST00000311487.5
ENST00000447654.1
ENST00000395004.3
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr12_-_10022735
|
0.531
|
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr10_-_105845674
|
0.445
|
ENST00000353479.5
ENST00000369733.3
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr12_+_32832203
|
0.399
|
ENST00000553257.1
ENST00000549701.1
ENST00000358214.5
ENST00000266481.6
ENST00000551476.1
ENST00000550154.1
ENST00000547312.1
ENST00000414834.2
ENST00000381000.4
ENST00000548750.1
|
DNM1L
|
dynamin 1-like
|
|
chr11_+_5710919
|
0.379
|
ENST00000379965.3
ENST00000425490.1
|
TRIM22
|
tripartite motif containing 22
|
|
chr11_-_102651343
|
0.373
|
ENST00000279441.4
ENST00000539681.1
|
MMP10
|
matrix metallopeptidase 10 (stromelysin 2)
|
|
chr2_+_90077680
|
0.345
|
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20
|
|
chr17_-_41466555
|
0.286
|
ENST00000586231.1
|
LINC00910
|
long intergenic non-protein coding RNA 910
|
|
chr1_+_155278539
|
0.277
|
ENST00000447866.1
|
FDPS
|
farnesyl diphosphate synthase
|
|
chr3_+_183853052
|
0.259
|
ENST00000273783.3
ENST00000432569.1
ENST00000444495.1
|
EIF2B5
|
eukaryotic translation initiation factor 2B, subunit 5 epsilon, 82kDa
|
|
chr6_-_111927062
|
0.255
|
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr1_+_155278625
|
0.250
|
ENST00000368356.4
ENST00000356657.6
|
FDPS
|
farnesyl diphosphate synthase
|
|
chr12_-_112279694
|
0.241
|
ENST00000443596.1
ENST00000442119.1
|
MAPKAPK5-AS1
|
MAPKAPK5 antisense RNA 1
|
|
chrX_+_43515467
|
0.236
|
ENST00000338702.3
ENST00000542639.1
|
MAOA
|
monoamine oxidase A
|
|
chr10_+_124914285
|
0.234
|
ENST00000407911.2
|
BUB3
|
BUB3 mitotic checkpoint protein
|
|
chr20_-_50722183
|
0.233
|
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein
|
|
chr2_+_217524323
|
0.219
|
ENST00000456764.1
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa
|
|
chrX_-_77225135
|
0.214
|
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4
|
|
chr11_-_59383617
|
0.199
|
ENST00000263847.1
|
OSBP
|
oxysterol binding protein
|
|
chr15_+_64680003
|
0.198
|
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4
|
|
chr6_+_26402517
|
0.195
|
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1
|
|
chr1_-_190446759
|
0.163
|
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3
|
|
chr8_-_102803163
|
0.163
|
ENST00000523645.1
ENST00000520346.1
ENST00000220931.6
ENST00000522448.1
ENST00000522951.1
ENST00000522252.1
ENST00000519098.1
|
NCALD
|
neurocalcin delta
|
|
chr19_+_48949030
|
0.163
|
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1
|
|
chr12_-_12837423
|
0.158
|
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19
|
|
chr1_-_67266939
|
0.152
|
ENST00000304526.2
|
INSL5
|
insulin-like 5
|
|
chr17_-_45266542
|
0.151
|
ENST00000531206.1
ENST00000527547.1
ENST00000446365.2
ENST00000575483.1
ENST00000066544.3
|
CDC27
|
cell division cycle 27
|
|
chr3_+_23851928
|
0.147
|
ENST00000467766.1
ENST00000424381.1
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1
|
|
chrX_+_30261847
|
0.146
|
ENST00000378981.3
ENST00000397550.1
|
MAGEB1
|
melanoma antigen family B, 1
|
|
chr14_+_103851712
|
0.145
|
ENST00000440884.3
ENST00000416682.2
ENST00000429436.2
ENST00000303622.9
|
MARK3
|
MAP/microtubule affinity-regulating kinase 3
|
|
chrX_+_44703249
|
0.144
|
ENST00000339042.4
|
DUSP21
|
dual specificity phosphatase 21
|
|
chrX_+_77154935
|
0.134
|
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb
|
|
chr6_-_94129244
|
0.128
|
ENST00000369303.4
ENST00000369297.1
|
EPHA7
|
EPH receptor A7
|
|
chr11_+_73498898
|
0.127
|
ENST00000535529.1
ENST00000497094.2
ENST00000411840.2
ENST00000535277.1
ENST00000398483.3
ENST00000542303.1
|
MRPL48
|
mitochondrial ribosomal protein L48
|
|
chr11_-_3859089
|
0.125
|
ENST00000396979.1
|
RHOG
|
ras homolog family member G
|
|
chr19_-_6424783
|
0.125
|
ENST00000398148.3
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr8_-_42623747
|
0.124
|
ENST00000534622.1
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6 (neuronal)
|
|
chr1_+_153747746
|
0.109
|
ENST00000368661.3
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3
|
|
chrX_+_135730373
|
0.109
|
ENST00000370628.2
|
CD40LG
|
CD40 ligand
|
|
chr1_+_186265399
|
0.108
|
ENST00000367486.3
ENST00000367484.3
ENST00000533951.1
ENST00000367482.4
ENST00000367483.4
ENST00000367485.4
ENST00000445192.2
|
PRG4
|
proteoglycan 4
|
|
chr6_-_111927449
|
0.108
|
ENST00000368761.5
ENST00000392556.4
ENST00000340026.6
|
TRAF3IP2
|
TRAF3 interacting protein 2
|
|
chr1_+_62439037
|
0.102
|
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila)
|
|
chr10_+_124913930
|
0.085
|
ENST00000368858.5
|
BUB3
|
BUB3 mitotic checkpoint protein
|
|
chrX_+_135618258
|
0.084
|
ENST00000440515.1
ENST00000456412.1
|
VGLL1
|
vestigial like 1 (Drosophila)
|
|
chr5_-_20575959
|
0.083
|
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2
|
|
chr8_+_105235572
|
0.083
|
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr1_+_100818156
|
0.078
|
ENST00000336454.3
|
CDC14A
|
cell division cycle 14A
|
|
chr9_+_125133315
|
0.074
|
ENST00000223423.4
ENST00000362012.2
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr1_+_153746683
|
0.073
|
ENST00000271857.2
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3
|
|
chrX_-_6453159
|
0.072
|
ENST00000381089.3
ENST00000398729.1
|
VCX3A
|
variable charge, X-linked 3A
|
|
chr17_-_40337470
|
0.072
|
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor
|
|
chr19_-_14785622
|
0.069
|
ENST00000443157.2
|
EMR3
|
egf-like module containing, mucin-like, hormone receptor-like 3
|
|
chr9_-_21202204
|
0.067
|
ENST00000239347.3
|
IFNA7
|
interferon, alpha 7
|
|
chr7_+_50348268
|
0.065
|
ENST00000438033.1
ENST00000439701.1
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr19_-_14785698
|
0.065
|
ENST00000344373.4
ENST00000595472.1
|
EMR3
|
egf-like module containing, mucin-like, hormone receptor-like 3
|
|
chr1_-_152386732
|
0.063
|
ENST00000271835.3
|
CRNN
|
cornulin
|
|
chr8_-_30670384
|
0.062
|
ENST00000221138.4
ENST00000518243.1
|
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme
|
|
chr2_+_27008865
|
0.062
|
ENST00000335756.4
ENST00000233505.8
|
CENPA
|
centromere protein A
|
|
chr1_-_211307404
|
0.061
|
ENST00000367007.4
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1
|
|
chr19_+_11071546
|
0.061
|
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
|
|
chr2_-_169887827
|
0.060
|
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11
|
|
chr2_+_166095898
|
0.060
|
ENST00000424833.1
ENST00000375437.2
ENST00000357398.3
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr22_+_22730353
|
0.060
|
ENST00000390296.2
|
IGLV5-45
|
immunoglobulin lambda variable 5-45
|
|
chr19_-_14785674
|
0.056
|
ENST00000253673.5
|
EMR3
|
egf-like module containing, mucin-like, hormone receptor-like 3
|
|
chr1_-_28969517
|
0.055
|
ENST00000263974.4
ENST00000373824.4
|
TAF12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa
|
|
chr9_-_21239978
|
0.055
|
ENST00000380222.2
|
IFNA14
|
interferon, alpha 14
|
|
chr4_-_66536196
|
0.049
|
ENST00000511294.1
|
EPHA5
|
EPH receptor A5
|
|
chr3_+_129247479
|
0.049
|
ENST00000296271.3
|
RHO
|
rhodopsin
|
|
chr3_+_186560462
|
0.046
|
ENST00000412955.2
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing
|
|
chr3_+_186560476
|
0.046
|
ENST00000320741.2
ENST00000444204.2
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing
|
|
chr2_-_89597542
|
0.046
|
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional)
|
|
chr9_-_21368075
|
0.044
|
ENST00000449498.1
|
IFNA13
|
interferon, alpha 13
|
|
chr6_+_30687978
|
0.043
|
ENST00000327892.8
ENST00000435534.1
|
TUBB
|
tubulin, beta class I
|
|
chrX_-_18690210
|
0.042
|
ENST00000379984.3
|
RS1
|
retinoschisin 1
|
|
chr4_+_80584903
|
0.040
|
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1
|
|
chr17_-_10372875
|
0.039
|
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr10_-_114206649
|
0.039
|
ENST00000369404.3
ENST00000369405.3
|
ZDHHC6
|
zinc finger, DHHC-type containing 6
|
|
chr14_+_32798547
|
0.038
|
ENST00000557354.1
ENST00000557102.1
ENST00000557272.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr1_+_28261492
|
0.034
|
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B
|
|
chr4_-_74486347
|
0.031
|
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6
|
|
chr6_+_154360357
|
0.030
|
ENST00000330432.7
ENST00000360422.4
|
OPRM1
|
opioid receptor, mu 1
|
|
chr1_+_100818009
|
0.030
|
ENST00000370125.2
ENST00000361544.6
ENST00000370124.3
|
CDC14A
|
cell division cycle 14A
|
|
chr4_+_110736659
|
0.030
|
ENST00000394631.3
ENST00000226796.6
|
GAR1
|
GAR1 ribonucleoprotein
|
|
chr12_-_25348007
|
0.028
|
ENST00000354189.5
ENST00000545133.1
ENST00000554347.1
ENST00000395987.3
ENST00000320267.9
ENST00000395990.2
ENST00000537577.1
|
CASC1
|
cancer susceptibility candidate 1
|
|
chr15_+_93443419
|
0.028
|
ENST00000557381.1
ENST00000420239.2
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr4_-_66536057
|
0.025
|
ENST00000273854.3
|
EPHA5
|
EPH receptor A5
|
|
chr16_-_20702578
|
0.021
|
ENST00000307493.4
ENST00000219151.4
|
ACSM1
|
acyl-CoA synthetase medium-chain family member 1
|
|
chr10_+_124913793
|
0.019
|
ENST00000368865.4
ENST00000538238.1
ENST00000368859.2
|
BUB3
|
BUB3 mitotic checkpoint protein
|
|
chr11_+_33061543
|
0.019
|
ENST00000432887.1
ENST00000528898.1
ENST00000531632.2
|
TCP11L1
|
t-complex 11, testis-specific-like 1
|
|
chr12_-_10959892
|
0.019
|
ENST00000240615.2
|
TAS2R8
|
taste receptor, type 2, member 8
|
|
chr10_+_18549645
|
0.019
|
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit
|
|
chr2_+_113763031
|
0.018
|
ENST00000259211.6
|
IL36A
|
interleukin 36, alpha
|
|
chr9_+_34646651
|
0.017
|
ENST00000378842.3
|
GALT
|
galactose-1-phosphate uridylyltransferase
|
|
chrX_-_8139308
|
0.016
|
ENST00000317103.4
|
VCX2
|
variable charge, X-linked 2
|
|
chr7_-_86849883
|
0.013
|
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial
|
|
chr1_-_151804314
|
0.012
|
ENST00000318247.6
|
RORC
|
RAR-related orphan receptor C
|
|
chr18_+_22040620
|
0.010
|
ENST00000426880.2
|
HRH4
|
histamine receptor H4
|
|
chr9_+_34646624
|
0.007
|
ENST00000450095.2
ENST00000556278.1
|
GALT
GALT
|
galactose-1-phosphate uridylyltransferase
Uncharacterized protein
|
|
chr20_-_1447467
|
0.003
|
ENST00000353088.2
ENST00000350991.4
|
NSFL1C
|
NSFL1 (p97) cofactor (p47)
|
|
chrX_-_13835147
|
0.002
|
ENST00000493677.1
ENST00000355135.2
|
GPM6B
|
glycoprotein M6B
|
|
chr1_+_192605252
|
0.002
|
ENST00000391995.2
ENST00000543215.1
|
RGS13
|
regulator of G-protein signaling 13
|
|
chr18_+_22040593
|
0.002
|
ENST00000256906.4
|
HRH4
|
histamine receptor H4
|
|
chr2_+_74682150
|
0.001
|
ENST00000233331.7
ENST00000431187.1
ENST00000409917.1
ENST00000409493.2
|
INO80B
|
INO80 complex subunit B
|