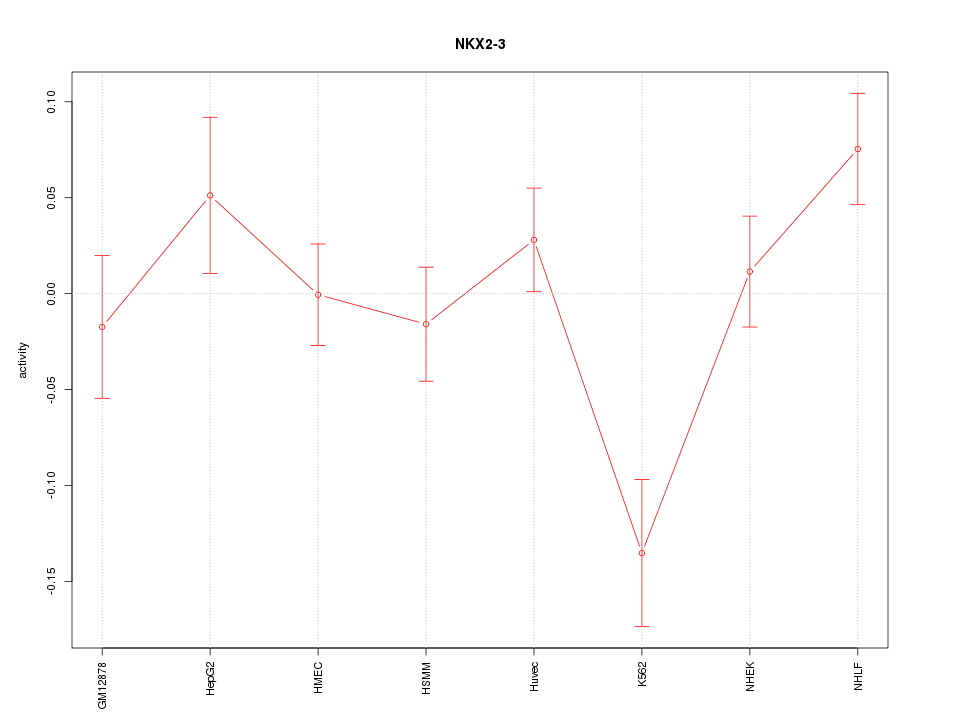
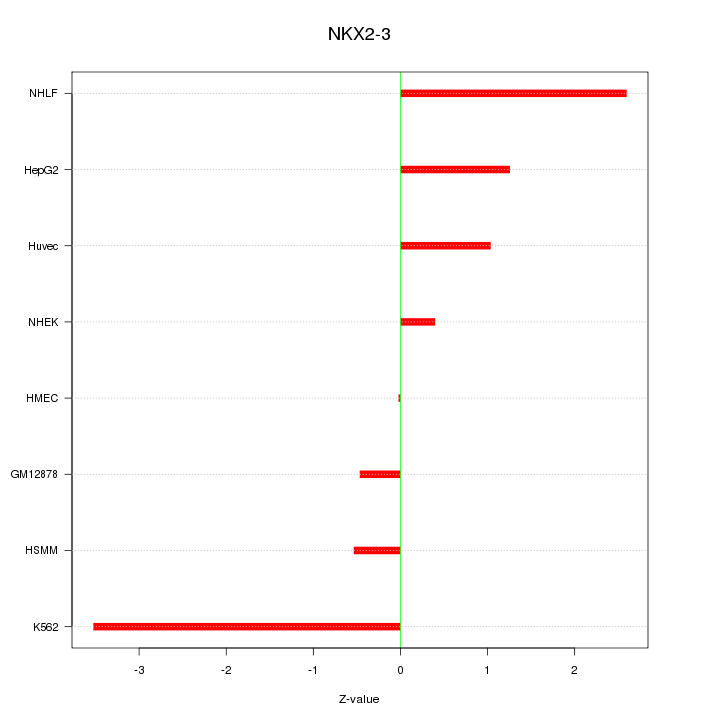
Motif ID: NKX2-3
Z-value: 1.677
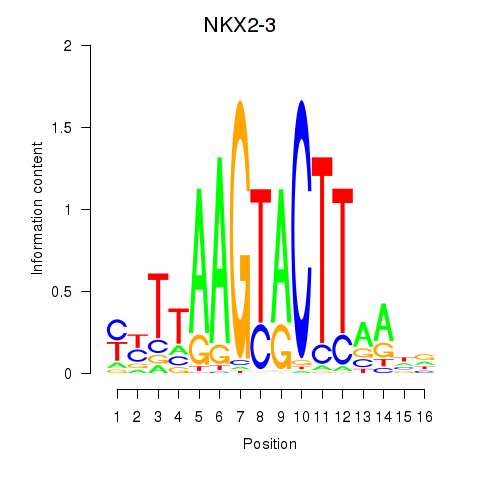
Transcription factors associated with NKX2-3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NKX2-3 | ENSG00000119919.9 | NKX2-3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 1.1 | 4.5 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.8 | 2.3 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.5 | 2.2 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.4 | 1.3 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.4 | 2.7 | GO:0060414 | aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 | 1.4 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.3 | 1.3 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.3 | 3.6 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.2 | 0.7 | GO:0014831 | intestine smooth muscle contraction(GO:0014827) gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.2 | 1.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 1.3 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.2 | 1.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.2 | 1.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 1.8 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.2 | 1.0 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.6 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 1.7 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 1.1 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.1 | 1.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 1.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.6 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 1.0 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.7 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 1.0 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.1 | 1.0 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.6 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.1 | 0.3 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 1.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.5 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.6 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 0.3 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.1 | 0.3 | GO:0010842 | retina layer formation(GO:0010842) camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 2.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.6 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 0.3 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 4.5 | GO:0030814 | regulation of cAMP metabolic process(GO:0030814) |
| 0.0 | 0.3 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.1 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) negative regulation of cell cycle G2/M phase transition(GO:1902750) |
| 0.0 | 0.3 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.1 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.0 | 0.6 | GO:0044550 | secondary metabolite biosynthetic process(GO:0044550) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.8 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.3 | GO:0048387 | protein maturation by protein folding(GO:0022417) negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 1.9 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.3 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 2.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.0 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.7 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0043628 | ncRNA 3'-end processing(GO:0043628) |
| 0.0 | 0.4 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 | 1.0 | GO:0046320 | regulation of fatty acid oxidation(GO:0046320) |
| 0.0 | 0.5 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.5 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.8 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.6 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.5 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.9 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 1.6 | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 2.1 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.4 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 1.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.1 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 5.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 4.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.3 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 11.4 | GO:0005625 | obsolete soluble fraction(GO:0005625) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.2 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 1.5 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 3.5 | GO:0005792 | obsolete microsome(GO:0005792) |
| 0.0 | 1.5 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.5 | 2.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 6.9 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.4 | 1.7 | GO:0019862 | IgA binding(GO:0019862) |
| 0.4 | 5.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 0.8 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.2 | 1.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 1.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.6 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.2 | 0.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 0.5 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 0.6 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 3.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.6 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.5 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.5 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.4 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.6 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 1.0 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.7 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) alpha-catenin binding(GO:0045294) |
| 0.1 | 1.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.7 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 1.0 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 2.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.6 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.1 | 1.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.2 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.1 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.2 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.6 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) |
| 0.0 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.6 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.8 | GO:0030552 | cyclic nucleotide binding(GO:0030551) cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) |
| 0.0 | 3.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.1 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.7 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |