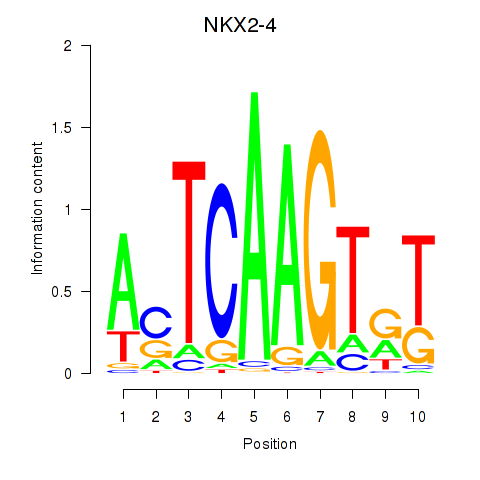
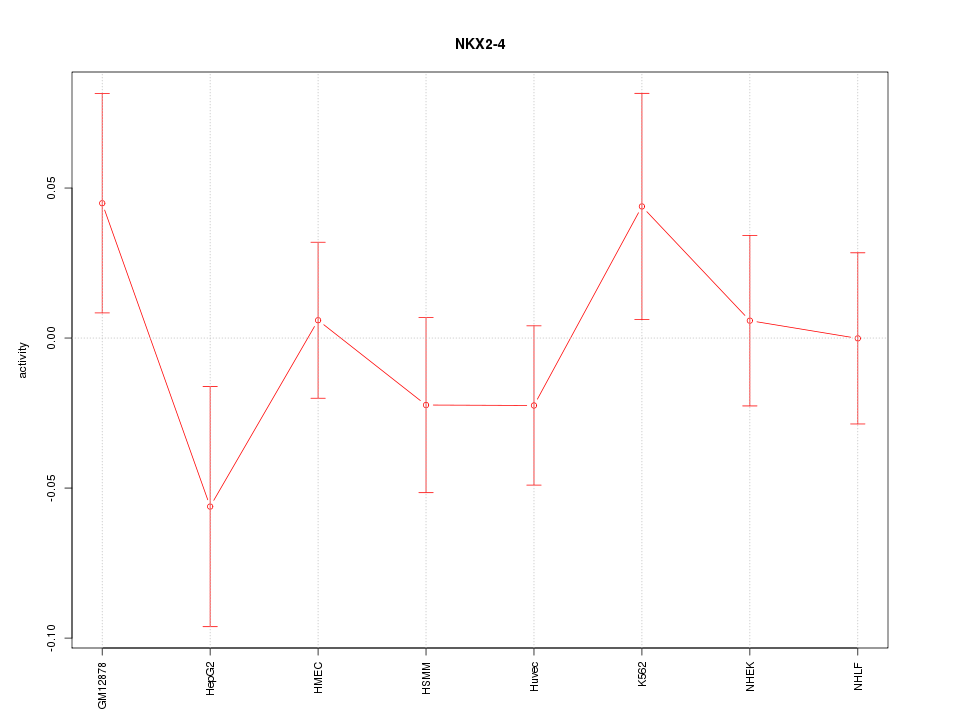
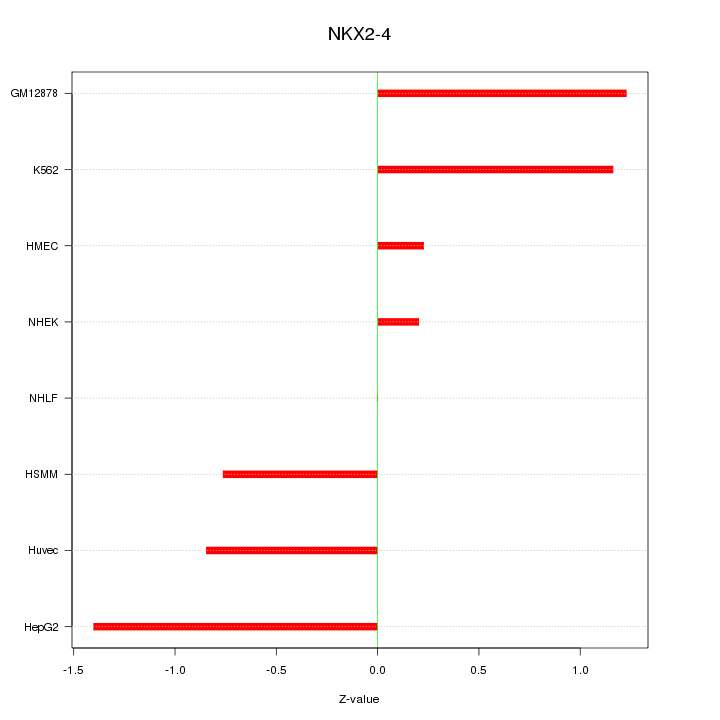
| 0.2 |
0.7 |
GO:0002901 |
mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) purine nucleotide salvage(GO:0032261) nucleotide salvage(GO:0043173) inosine metabolic process(GO:0046102) xanthine metabolic process(GO:0046110) |
| 0.2 |
0.6 |
GO:0002541 |
activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.2 |
1.0 |
GO:0043316 |
natural killer cell activation involved in immune response(GO:0002323) cytotoxic T cell degranulation(GO:0043316) natural killer cell degranulation(GO:0043320) |
| 0.2 |
0.5 |
GO:0043366 |
beta selection(GO:0043366) |
| 0.2 |
0.5 |
GO:0097237 |
response to cobalt ion(GO:0032025) cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 |
0.4 |
GO:0006478 |
peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 |
0.4 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 |
0.5 |
GO:0044854 |
membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) positive regulation of dopamine receptor signaling pathway(GO:0060161) caveola assembly(GO:0070836) membrane assembly(GO:0071709) |
| 0.1 |
0.5 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 |
0.5 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.1 |
0.4 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.1 |
0.4 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.1 |
0.4 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 |
0.4 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 |
0.4 |
GO:0008588 |
release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 |
0.4 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.1 |
0.8 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 |
0.3 |
GO:1902414 |
protein localization to adherens junction(GO:0071896) protein localization to cell junction(GO:1902414) |
| 0.1 |
0.2 |
GO:0045938 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 |
0.4 |
GO:0043615 |
astrocyte cell migration(GO:0043615) |
| 0.1 |
0.2 |
GO:0018243 |
protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 |
0.6 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.1 |
0.7 |
GO:0020027 |
hemoglobin metabolic process(GO:0020027) |
| 0.1 |
0.3 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.0 |
0.1 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.0 |
0.3 |
GO:0060219 |
camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 |
0.2 |
GO:0046037 |
GMP metabolic process(GO:0046037) |
| 0.0 |
0.3 |
GO:0007499 |
ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 |
0.7 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.1 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.0 |
0.4 |
GO:0015705 |
iodide transport(GO:0015705) |
| 0.0 |
0.1 |
GO:0050942 |
positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 |
0.2 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.0 |
0.2 |
GO:0006122 |
mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 |
0.2 |
GO:0031580 |
membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 |
0.1 |
GO:0019056 |
modulation by virus of host transcription(GO:0019056) regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 |
0.2 |
GO:0042866 |
pyruvate biosynthetic process(GO:0042866) |
| 0.0 |
0.4 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.1 |
GO:1904037 |
positive regulation of epithelial cell apoptotic process(GO:1904037) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 |
0.5 |
GO:0048268 |
negative regulation of receptor-mediated endocytosis(GO:0048261) clathrin coat assembly(GO:0048268) |
| 0.0 |
0.5 |
GO:0007128 |
meiotic prophase I(GO:0007128) |
| 0.0 |
0.3 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.2 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.0 |
0.1 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.0 |
0.3 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 |
1.9 |
GO:0006821 |
chloride transport(GO:0006821) |
| 0.0 |
0.1 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 |
0.2 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.0 |
0.8 |
GO:0033209 |
tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 |
0.6 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.0 |
0.3 |
GO:0033261 |
obsolete regulation of S phase(GO:0033261) |
| 0.0 |
0.2 |
GO:0070189 |
kynurenine metabolic process(GO:0070189) |
| 0.0 |
0.2 |
GO:1901984 |
negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 |
0.4 |
GO:0030252 |
growth hormone secretion(GO:0030252) |
| 0.0 |
0.1 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 |
0.1 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.0 |
0.5 |
GO:0050892 |
intestinal absorption(GO:0050892) |
| 0.0 |
0.1 |
GO:0015722 |
canalicular bile acid transport(GO:0015722) |
| 0.0 |
0.3 |
GO:0007096 |
regulation of exit from mitosis(GO:0007096) |
| 0.0 |
0.1 |
GO:0045003 |
DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 |
0.1 |
GO:2000008 |
regulation of protein localization to cell surface(GO:2000008) |
| 0.0 |
0.1 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 |
0.2 |
GO:0060750 |
epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 |
2.4 |
GO:0042113 |
B cell activation(GO:0042113) |
| 0.0 |
0.1 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
1.5 |
GO:0007229 |
integrin-mediated signaling pathway(GO:0007229) |
| 0.0 |
0.1 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.1 |
GO:0010988 |
regulation of low-density lipoprotein particle clearance(GO:0010988) regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 |
0.4 |
GO:0006611 |
protein export from nucleus(GO:0006611) |
| 0.0 |
0.1 |
GO:0031282 |
regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 |
0.3 |
GO:0032784 |
regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 |
0.1 |
GO:0030422 |
production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 |
0.1 |
GO:0006069 |
ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 |
0.1 |
GO:0003416 |
endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 |
0.1 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 |
0.1 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.0 |
0.0 |
GO:0006530 |
asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 |
0.0 |
GO:0001552 |
ovarian follicle atresia(GO:0001552) |
| 0.0 |
0.1 |
GO:0006562 |
proline catabolic process(GO:0006562) |
| 0.0 |
0.3 |
GO:0002026 |
regulation of the force of heart contraction(GO:0002026) |
| 0.0 |
0.3 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.0 |
GO:0010919 |
regulation of inositol phosphate biosynthetic process(GO:0010919) positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 |
0.1 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.1 |
GO:0002921 |
negative regulation of humoral immune response(GO:0002921) negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 |
0.1 |
GO:0090080 |
positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 |
0.1 |
GO:0006379 |
mRNA cleavage(GO:0006379) |