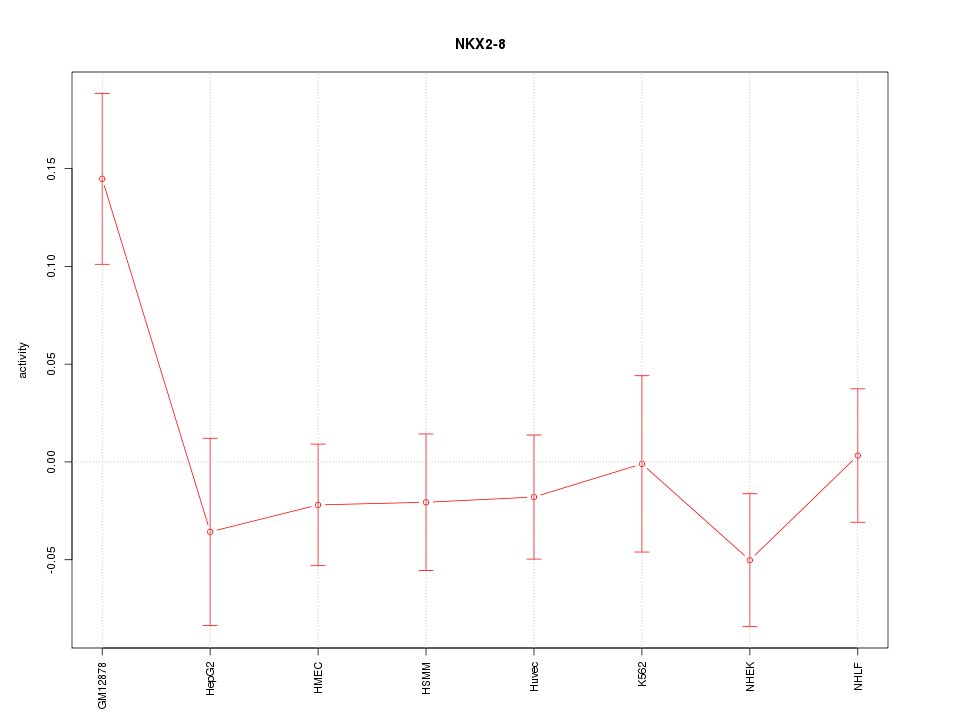
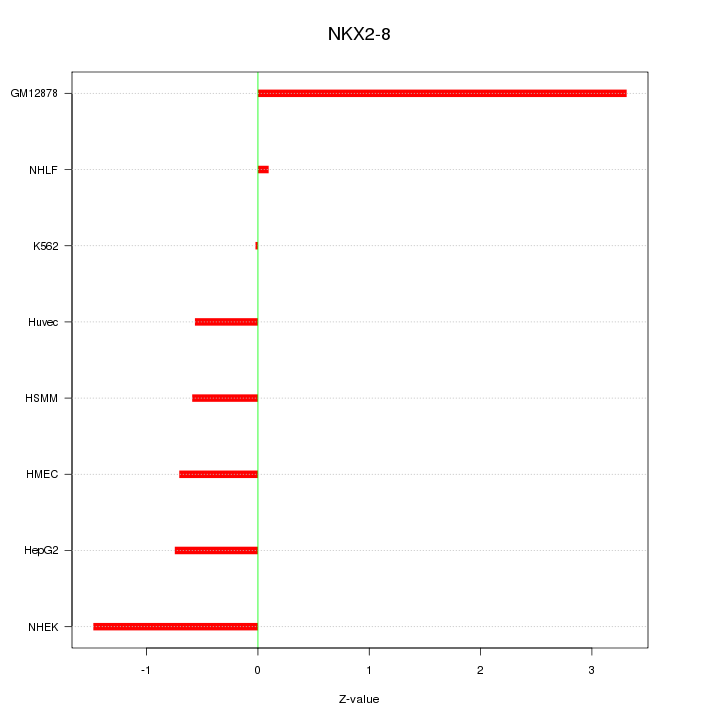
Motif ID: NKX2-8
Z-value: 1.364
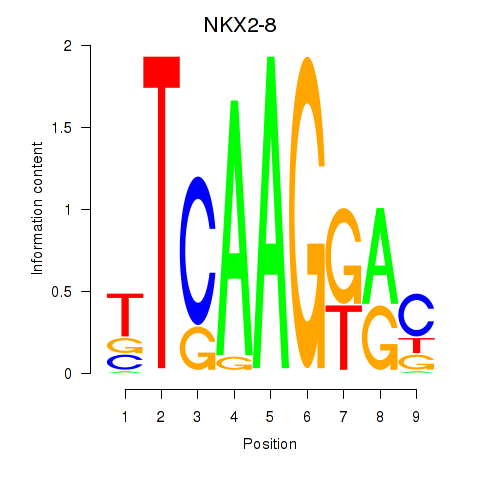
Transcription factors associated with NKX2-8:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NKX2-8 | ENSG00000136327.6 | NKX2-8 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.4 | 6.5 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.3 | 0.8 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.2 | 0.7 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.2 | 1.9 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.1 | 0.6 | GO:0045938 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.1 | 0.6 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.8 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 1.4 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.4 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.1 | 0.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 1.9 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 0.2 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.1 | 0.2 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.9 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.1 | 2.5 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.1 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 1.7 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.2 | GO:0030578 | nuclear body organization(GO:0030575) PML body organization(GO:0030578) |
| 0.0 | 0.3 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0001807 | type IV hypersensitivity(GO:0001806) regulation of type IV hypersensitivity(GO:0001807) |
| 0.0 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.3 | GO:1904893 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.3 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.8 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.0 | 1.2 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.3 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.4 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.5 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0001974 | blood vessel remodeling(GO:0001974) |
| 0.0 | 0.0 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.0 | 0.3 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 0.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 1.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.7 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.3 | 0.8 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.2 | 0.9 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 1.5 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 0.5 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.1 | 1.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.7 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.8 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.8 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.6 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 0.2 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.3 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 2.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.0 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0050833 | pyruvate secondary active transmembrane transporter activity(GO:0005477) pyruvate transmembrane transporter activity(GO:0050833) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.9 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.7 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.5 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |