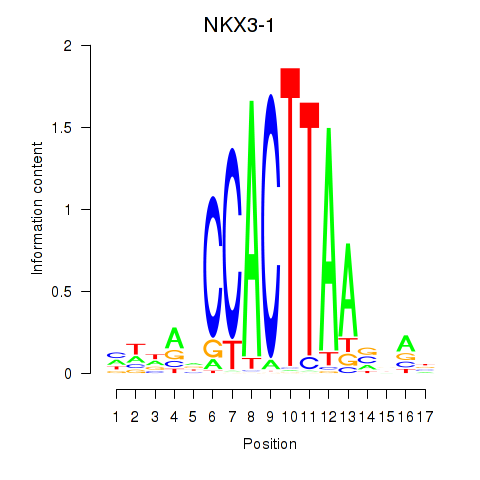
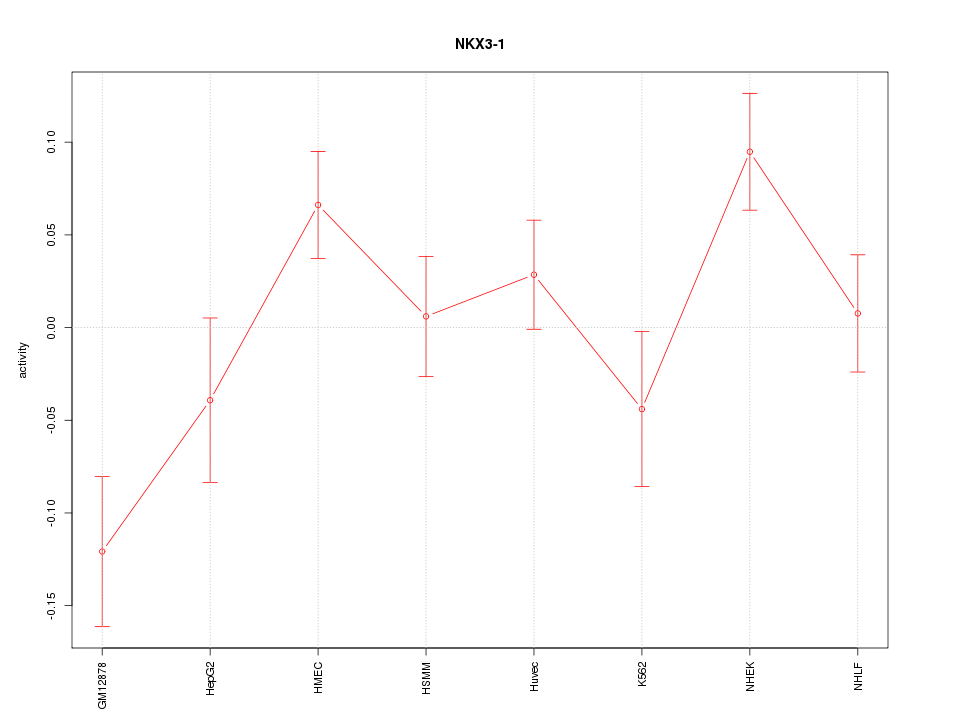
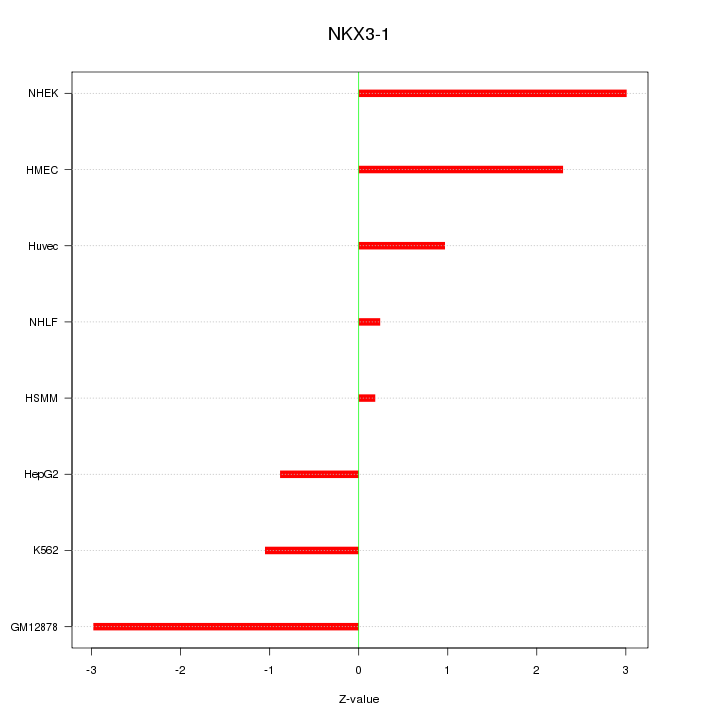
| 1.1 |
6.3 |
GO:0007499 |
ectoderm and mesoderm interaction(GO:0007499) |
| 0.6 |
2.8 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.5 |
2.1 |
GO:0006663 |
platelet activating factor biosynthetic process(GO:0006663) positive regulation of vesicle fusion(GO:0031340) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.5 |
3.1 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.4 |
2.8 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.3 |
1.9 |
GO:0060242 |
contact inhibition(GO:0060242) |
| 0.2 |
1.2 |
GO:0021535 |
cell migration in hindbrain(GO:0021535) |
| 0.2 |
0.7 |
GO:0019046 |
release from viral latency(GO:0019046) |
| 0.1 |
0.3 |
GO:0048670 |
regulation of collateral sprouting(GO:0048670) |
| 0.1 |
0.4 |
GO:0007028 |
cytoplasm organization(GO:0007028) ribonucleoprotein complex disassembly(GO:0032988) |
| 0.1 |
2.1 |
GO:0060391 |
positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 |
0.4 |
GO:0006982 |
response to lipid hydroperoxide(GO:0006982) |
| 0.1 |
0.7 |
GO:0006011 |
UDP-glucose metabolic process(GO:0006011) UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 |
0.4 |
GO:0009609 |
response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.1 |
1.0 |
GO:0019885 |
antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 |
0.6 |
GO:0022614 |
membrane to membrane docking(GO:0022614) |
| 0.1 |
6.7 |
GO:0030521 |
androgen receptor signaling pathway(GO:0030521) |
| 0.1 |
3.4 |
GO:0030837 |
negative regulation of actin filament polymerization(GO:0030837) |
| 0.1 |
0.3 |
GO:0051877 |
pigment granule aggregation in cell center(GO:0051877) |
| 0.1 |
1.7 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.1 |
0.4 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.9 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.6 |
GO:0097031 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
2.4 |
GO:0007193 |
adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 |
0.2 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.3 |
GO:0019614 |
phenol-containing compound catabolic process(GO:0019336) catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 |
0.3 |
GO:0044818 |
mitotic G2/M transition checkpoint(GO:0044818) protein neddylation(GO:0045116) |
| 0.0 |
0.3 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.0 |
0.1 |
GO:0051958 |
methotrexate transport(GO:0051958) |
| 0.0 |
0.2 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 |
1.6 |
GO:0006308 |
DNA catabolic process(GO:0006308) |
| 0.0 |
0.5 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.0 |
0.2 |
GO:0006346 |
methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 |
0.5 |
GO:0007128 |
meiotic prophase I(GO:0007128) |
| 0.0 |
0.5 |
GO:0019369 |
arachidonic acid metabolic process(GO:0019369) |
| 0.0 |
0.6 |
GO:0007271 |
synaptic transmission, cholinergic(GO:0007271) |
| 0.0 |
2.3 |
GO:0007229 |
integrin-mediated signaling pathway(GO:0007229) |
| 0.0 |
0.4 |
GO:0042403 |
thyroid hormone metabolic process(GO:0042403) |
| 0.0 |
0.1 |
GO:1902414 |
protein localization to adherens junction(GO:0071896) protein localization to cell junction(GO:1902414) |
| 0.0 |
0.7 |
GO:0006887 |
exocytosis(GO:0006887) |
| 0.0 |
0.3 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.3 |
GO:0021513 |
spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.0 |
0.8 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.5 |
GO:0048168 |
regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 |
0.4 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
2.2 |
GO:0010951 |
negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 |
0.7 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.3 |
GO:0034199 |
activation of protein kinase A activity(GO:0034199) |
| 0.0 |
0.5 |
GO:0042733 |
embryonic digit morphogenesis(GO:0042733) |
| 0.0 |
0.3 |
GO:0019933 |
cAMP-mediated signaling(GO:0019933) |
| 0.0 |
0.1 |
GO:0002862 |
negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 |
0.1 |
GO:0006122 |
mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 |
0.2 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.0 |
1.1 |
GO:0030814 |
regulation of cAMP metabolic process(GO:0030814) |
| 0.0 |
0.1 |
GO:0042693 |
muscle cell fate commitment(GO:0042693) |
| 0.0 |
0.1 |
GO:0071478 |
cellular response to radiation(GO:0071478) |
| 0.0 |
0.1 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
0.5 |
GO:0032480 |
negative regulation of type I interferon production(GO:0032480) |
| 0.0 |
0.8 |
GO:0006921 |
cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.0 |
2.1 |
GO:0006470 |
protein dephosphorylation(GO:0006470) |
| 0.0 |
0.3 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |