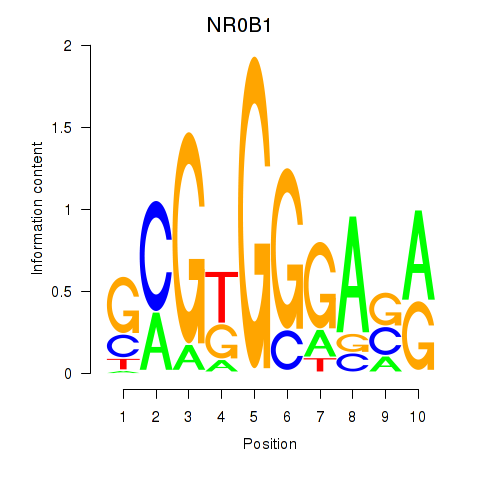
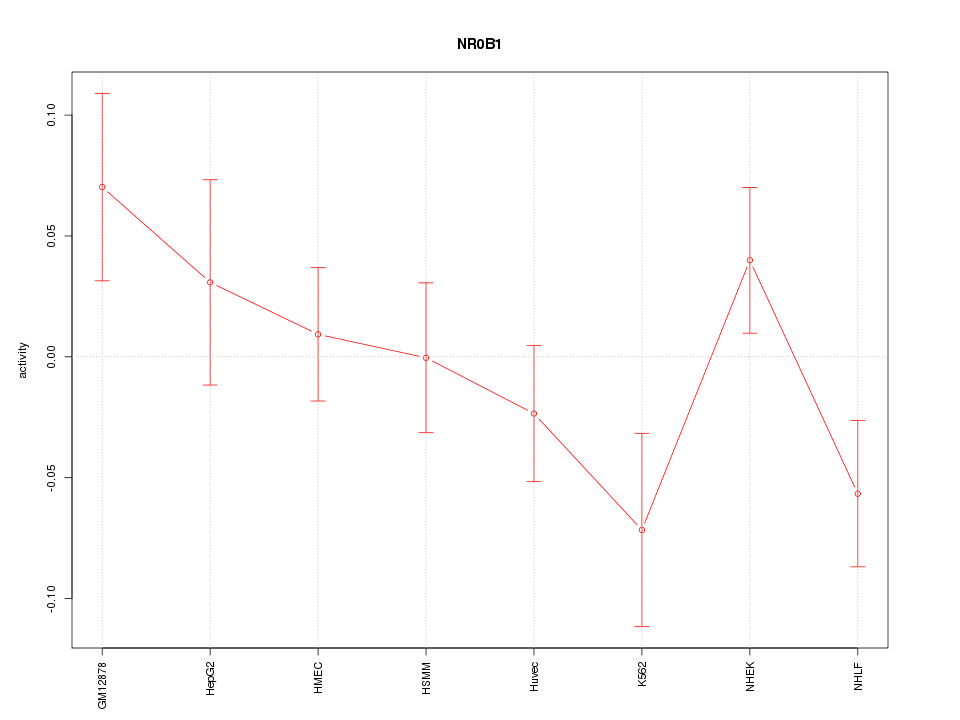
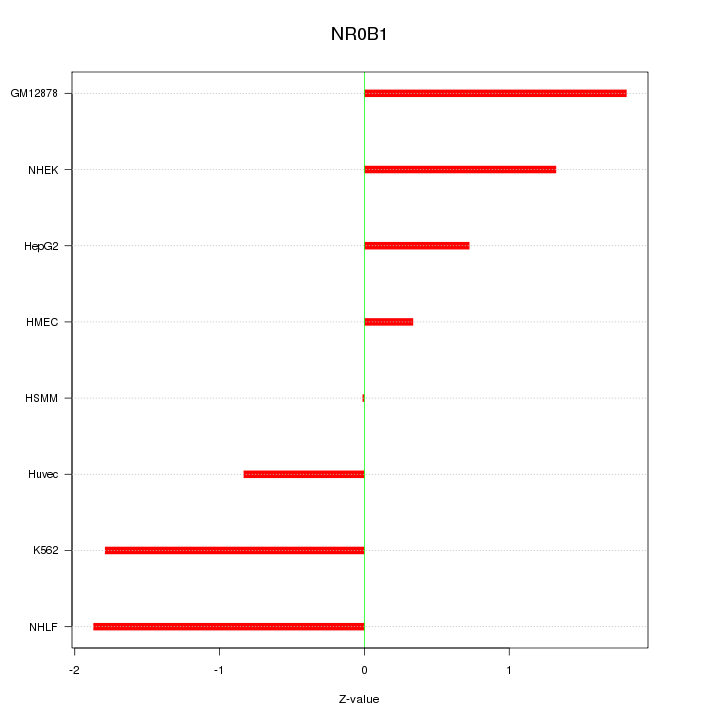
| 0.5 |
2.8 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.4 |
1.6 |
GO:0033602 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of dopamine secretion(GO:0033602) |
| 0.3 |
1.0 |
GO:0042097 |
interleukin-4 biosynthetic process(GO:0042097) positive regulation of interleukin-10 biosynthetic process(GO:0045082) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.3 |
1.9 |
GO:0002335 |
mature B cell differentiation(GO:0002335) |
| 0.2 |
0.7 |
GO:0030205 |
dermatan sulfate metabolic process(GO:0030205) dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 |
1.9 |
GO:0051552 |
flavone metabolic process(GO:0051552) |
| 0.2 |
0.5 |
GO:0006382 |
adenosine to inosine editing(GO:0006382) |
| 0.1 |
0.3 |
GO:0060054 |
positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 |
0.4 |
GO:0014887 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 |
0.4 |
GO:0021896 |
forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.1 |
0.4 |
GO:0010837 |
regulation of keratinocyte proliferation(GO:0010837) |
| 0.1 |
0.9 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.1 |
0.3 |
GO:0003011 |
diaphragm contraction(GO:0002086) involuntary skeletal muscle contraction(GO:0003011) |
| 0.1 |
3.8 |
GO:0016925 |
protein sumoylation(GO:0016925) |
| 0.1 |
1.7 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.1 |
0.3 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.1 |
1.0 |
GO:0051005 |
negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 |
0.4 |
GO:0060528 |
secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 |
0.8 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.1 |
0.2 |
GO:0033364 |
endosome transport via multivesicular body sorting pathway(GO:0032509) endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mast cell secretory granule organization(GO:0033364) |
| 0.1 |
0.5 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.1 |
0.1 |
GO:1901724 |
positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.1 |
2.7 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.1 |
0.4 |
GO:0006562 |
proline catabolic process(GO:0006562) |
| 0.1 |
0.7 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.1 |
0.5 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) |
| 0.1 |
0.2 |
GO:0034587 |
RNA 5'-end processing(GO:0000966) piRNA metabolic process(GO:0034587) |
| 0.1 |
0.2 |
GO:0043116 |
negative regulation of vascular permeability(GO:0043116) |
| 0.1 |
0.5 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 |
0.2 |
GO:0042816 |
pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 |
0.1 |
GO:0046532 |
regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 |
0.2 |
GO:0043276 |
anoikis(GO:0043276) |
| 0.0 |
0.2 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 |
0.2 |
GO:0006896 |
Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 |
0.4 |
GO:0060123 |
regulation of growth hormone secretion(GO:0060123) |
| 0.0 |
0.2 |
GO:0010832 |
negative regulation of myotube differentiation(GO:0010832) |
| 0.0 |
0.2 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 |
0.4 |
GO:0006998 |
nuclear envelope organization(GO:0006998) |
| 0.0 |
0.3 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 |
0.2 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.0 |
0.1 |
GO:0000212 |
meiotic spindle organization(GO:0000212) |
| 0.0 |
0.1 |
GO:0070734 |
histone H3-K27 methylation(GO:0070734) |
| 0.0 |
0.1 |
GO:0048852 |
diencephalon morphogenesis(GO:0048852) |
| 0.0 |
0.1 |
GO:0048073 |
negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) regulation of eye pigmentation(GO:0048073) |
| 0.0 |
0.2 |
GO:0045945 |
positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 |
0.2 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.0 |
1.3 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.0 |
0.2 |
GO:0045060 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 |
0.1 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 |
0.2 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 |
0.1 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.0 |
0.2 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.0 |
0.1 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.6 |
GO:0050909 |
sensory perception of taste(GO:0050909) |
| 0.0 |
0.6 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 |
0.1 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 |
0.6 |
GO:0035050 |
embryonic heart tube development(GO:0035050) |
| 0.0 |
0.1 |
GO:0015670 |
carbon dioxide transport(GO:0015670) |
| 0.0 |
0.1 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.1 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 |
0.1 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.0 |
0.1 |
GO:0010799 |
regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 |
0.1 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 |
0.2 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.2 |
GO:0070534 |
protein K63-linked ubiquitination(GO:0070534) |
| 0.0 |
0.0 |
GO:0071603 |
myoblast migration(GO:0051451) endothelial cell-cell adhesion(GO:0071603) |
| 0.0 |
0.0 |
GO:0003099 |
positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 |
0.0 |
GO:0042986 |
positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 |
0.3 |
GO:0030838 |
positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 |
0.3 |
GO:0045773 |
positive regulation of axon extension(GO:0045773) |
| 0.0 |
0.2 |
GO:0015701 |
bicarbonate transport(GO:0015701) |