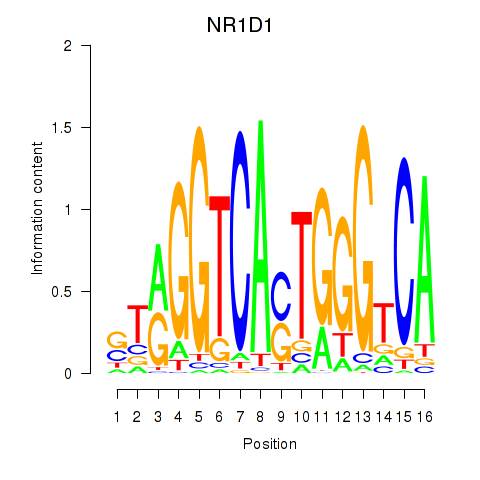
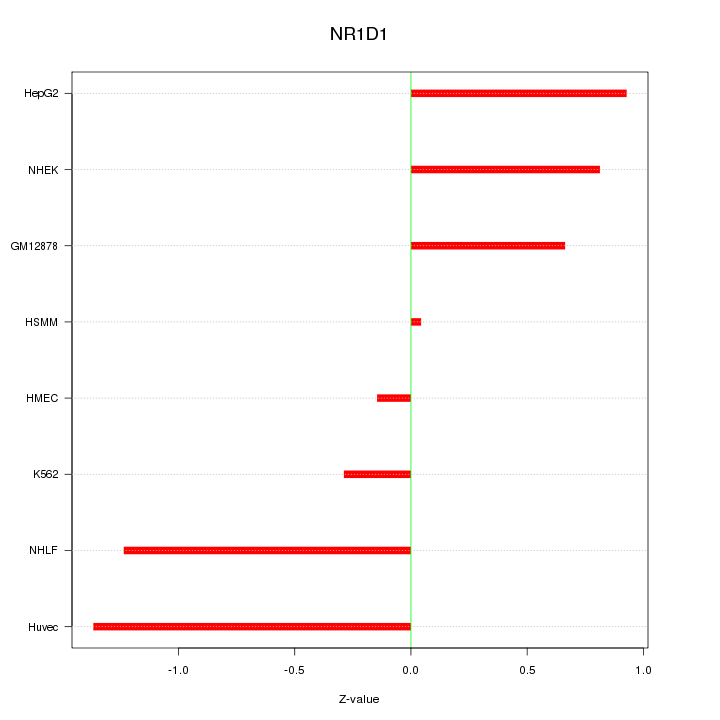
| 0.3 |
0.8 |
GO:0003289 |
septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) |
| 0.2 |
0.9 |
GO:0007262 |
STAT protein import into nucleus(GO:0007262) |
| 0.2 |
0.6 |
GO:0034139 |
regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.2 |
0.5 |
GO:0060557 |
positive regulation of chronic inflammatory response(GO:0002678) regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.2 |
0.5 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.1 |
0.6 |
GO:0009439 |
cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.1 |
0.3 |
GO:0032223 |
negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 |
0.1 |
GO:0032196 |
transposition, DNA-mediated(GO:0006313) transposition(GO:0032196) |
| 0.1 |
0.7 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
0.8 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 |
0.2 |
GO:0033197 |
response to vitamin E(GO:0033197) |
| 0.0 |
0.3 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.0 |
0.1 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 |
0.1 |
GO:0034122 |
negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 |
0.2 |
GO:0046642 |
negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 |
0.3 |
GO:0021978 |
telencephalon regionalization(GO:0021978) |
| 0.0 |
0.3 |
GO:0006108 |
malate metabolic process(GO:0006108) NADH metabolic process(GO:0006734) |
| 0.0 |
0.2 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 |
0.1 |
GO:0045726 |
negative regulation of macrophage cytokine production(GO:0010936) regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 |
0.3 |
GO:0046655 |
folic acid metabolic process(GO:0046655) |
| 0.0 |
0.1 |
GO:0015904 |
tetracycline transport(GO:0015904) |
| 0.0 |
0.8 |
GO:0046324 |
regulation of glucose import(GO:0046324) |
| 0.0 |
0.1 |
GO:0060528 |
secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 |
0.4 |
GO:0007405 |
neuroblast proliferation(GO:0007405) |