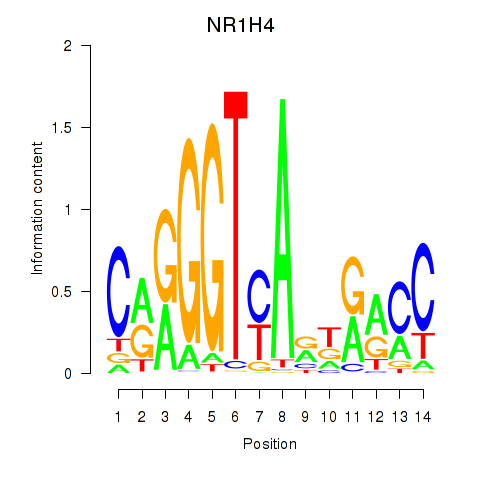
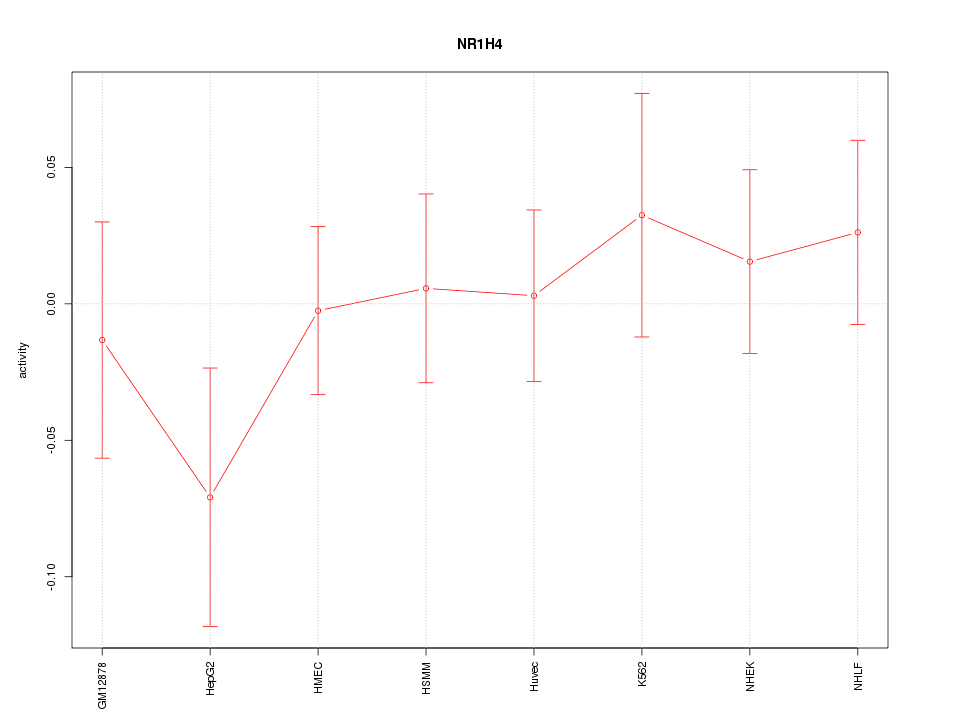
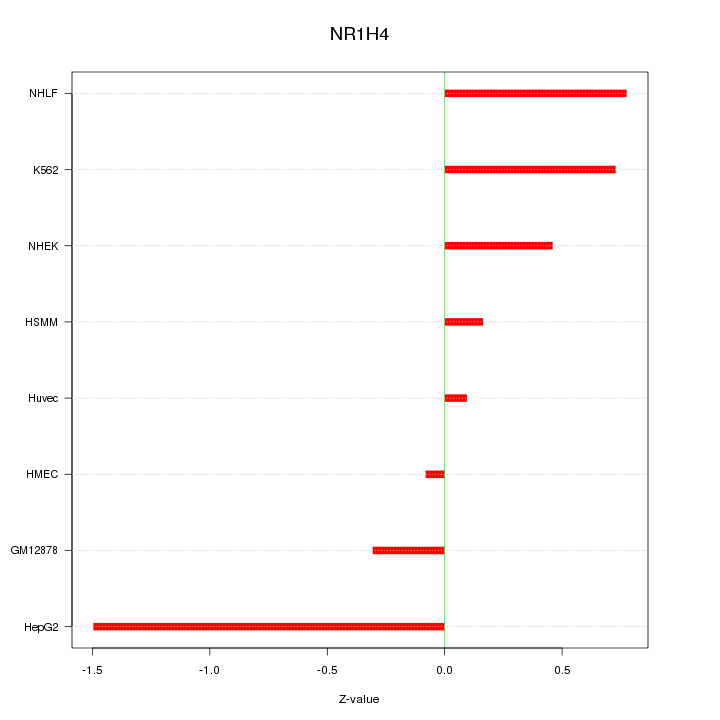
| 0.3 |
0.9 |
GO:0046604 |
regulation of mitotic centrosome separation(GO:0046602) positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.3 |
2.4 |
GO:0071692 |
sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.2 |
0.5 |
GO:0035408 |
histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 |
0.4 |
GO:0002158 |
osteoclast proliferation(GO:0002158) |
| 0.1 |
0.2 |
GO:0090598 |
male genitalia morphogenesis(GO:0048808) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 |
0.9 |
GO:0055015 |
ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 |
0.9 |
GO:0060638 |
mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.1 |
0.3 |
GO:0060710 |
chorio-allantoic fusion(GO:0060710) |
| 0.1 |
0.3 |
GO:0033632 |
regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 |
0.2 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.1 |
GO:0090086 |
negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 |
0.3 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.0 |
0.2 |
GO:0060414 |
aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 |
0.6 |
GO:0071230 |
positive regulation of TOR signaling(GO:0032008) cellular response to amino acid stimulus(GO:0071230) |
| 0.0 |
1.4 |
GO:0043462 |
regulation of ATPase activity(GO:0043462) |
| 0.0 |
0.2 |
GO:0034337 |
RNA folding(GO:0034337) DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) regulation of DNA methylation(GO:0044030) |
| 0.0 |
0.1 |
GO:0007196 |
adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 |
0.2 |
GO:0015824 |
proline transport(GO:0015824) |
| 0.0 |
0.3 |
GO:0002092 |
positive regulation of receptor internalization(GO:0002092) |
| 0.0 |
0.2 |
GO:0006772 |
thiamine metabolic process(GO:0006772) |
| 0.0 |
0.2 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.1 |
GO:0048263 |
determination of dorsal identity(GO:0048263) |
| 0.0 |
0.4 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.0 |
0.1 |
GO:0006922 |
obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 |
0.3 |
GO:0097031 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.2 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 |
0.1 |
GO:0060261 |
positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 |
0.2 |
GO:0019054 |
modulation by virus of host process(GO:0019054) modulation by symbiont of host cellular process(GO:0044068) |
| 0.0 |
0.1 |
GO:0003310 |
pancreatic A cell differentiation(GO:0003310) |
| 0.0 |
0.0 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 |
0.2 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.2 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.0 |
0.1 |
GO:0001507 |
acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 |
0.3 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.0 |
0.0 |
GO:0006122 |
mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |