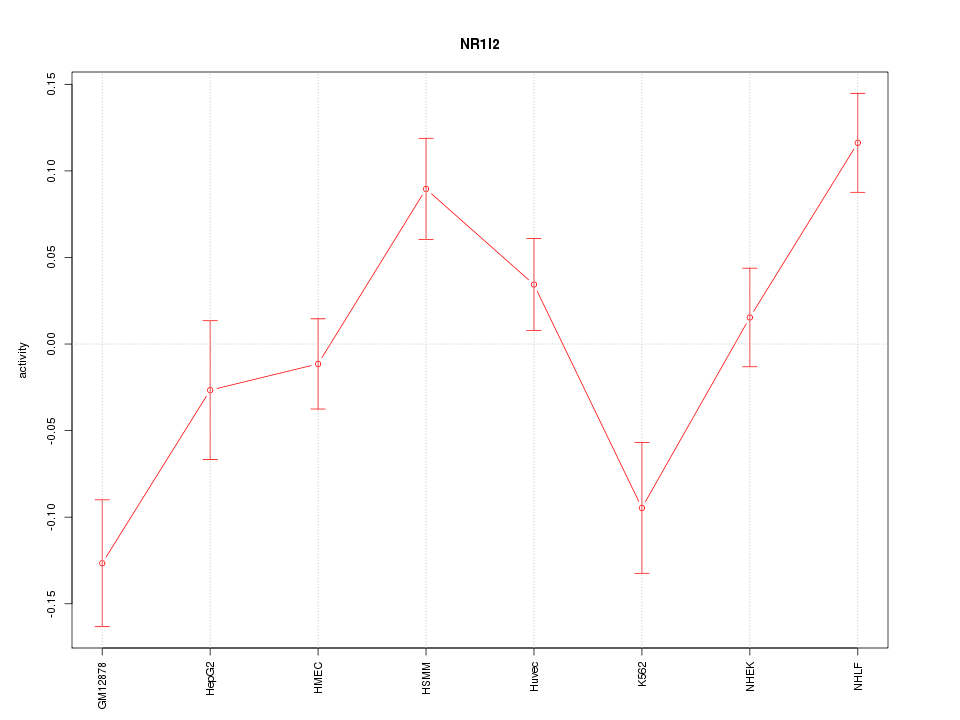
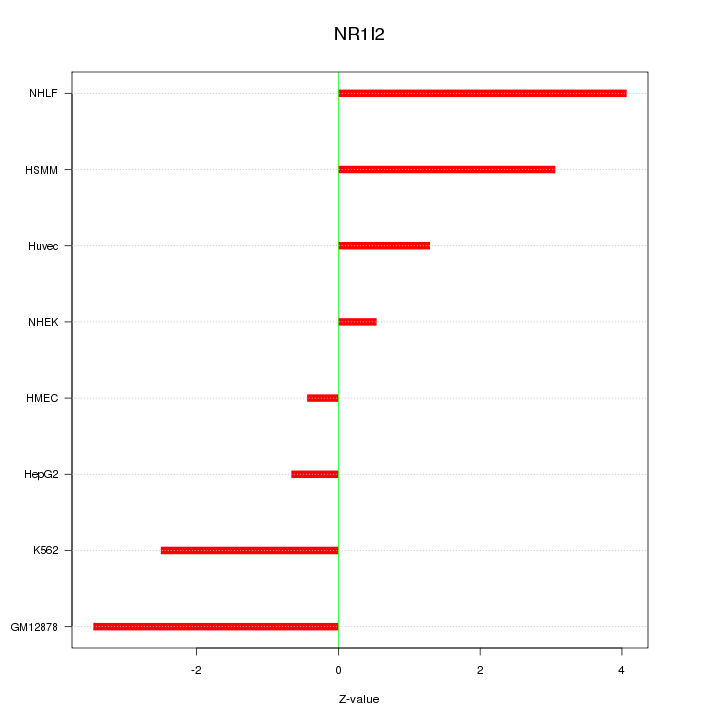
Motif ID: NR1I2
Z-value: 2.417
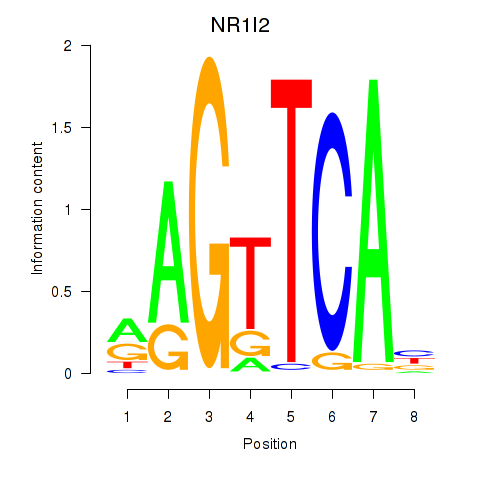
Transcription factors associated with NR1I2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| NR1I2 | ENSG00000144852.12 | NR1I2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 1.7 | 5.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 1.2 | 4.8 | GO:0009956 | radial pattern formation(GO:0009956) |
| 1.2 | 13.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.9 | 5.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.5 | 7.8 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.4 | 2.5 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.4 | 1.5 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.4 | 3.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.3 | 2.7 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.3 | 0.9 | GO:0002384 | hepatic immune response(GO:0002384) positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.3 | 2.8 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.3 | 1.3 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 | 0.4 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.2 | 4.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.2 | 0.5 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.2 | 1.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.2 | 0.5 | GO:0061117 | cardiac left ventricle formation(GO:0003218) negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.2 | 2.9 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 0.6 | GO:0043501 | skeletal muscle adaptation(GO:0043501) |
| 0.2 | 0.5 | GO:0042759 | fatty acid elongation, saturated fatty acid(GO:0019367) long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 1.2 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 | 0.3 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.1 | 0.7 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) regulation of cholesterol transporter activity(GO:0060694) |
| 0.1 | 0.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.6 | GO:0002331 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.7 | GO:0006789 | bilirubin conjugation(GO:0006789) biphenyl metabolic process(GO:0018879) biphenyl catabolic process(GO:0070980) |
| 0.1 | 2.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.4 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.1 | 0.8 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 0.4 | GO:0045760 | mechanosensory behavior(GO:0007638) response to auditory stimulus(GO:0010996) operant conditioning(GO:0035106) positive regulation of action potential(GO:0045760) |
| 0.1 | 0.6 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 0.3 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 0.5 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.1 | 0.4 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 1.0 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 3.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.3 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 0.5 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 6.3 | GO:0032387 | negative regulation of intracellular transport(GO:0032387) |
| 0.1 | 0.7 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.7 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 | 0.2 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.9 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.1 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 18.2 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.4 | GO:0051174 | regulation of phosphorus metabolic process(GO:0051174) |
| 0.0 | 0.9 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.5 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.6 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.5 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.2 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.7 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 1.2 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.5 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 2.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.0 | GO:0090281 | negative regulation of calcium ion import(GO:0090281) |
| 0.0 | 0.4 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.7 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.2 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.0 | 0.2 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.3 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 1.3 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 1.2 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.9 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.7 | GO:0035303 | regulation of dephosphorylation(GO:0035303) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.7 | GO:0090263 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of canonical Wnt signaling pathway(GO:0090263) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 1.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.3 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 2.1 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.6 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 1.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.9 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine production(GO:0032682) negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 1.2 | GO:0001906 | cell killing(GO:0001906) |
| 0.0 | 0.8 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 1.3 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 1.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.6 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 2.4 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.7 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 3.3 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 1.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.9 | 13.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.5 | 5.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 4.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.3 | 2.7 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 2.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 0.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 1.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 1.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 2.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 3.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 3.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 2.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.3 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 1.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.9 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 3.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 1.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.6 | GO:0001725 | stress fiber(GO:0001725) actin filament(GO:0005884) contractile actin filament bundle(GO:0097517) |
| 0.0 | 4.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 3.5 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.2 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 2.0 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.0 | 0.9 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.0 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.7 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.3 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.0 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.8 | 2.5 | GO:0048184 | obsolete follistatin binding(GO:0048184) |
| 0.6 | 8.5 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.5 | 6.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 12.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 3.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.3 | 2.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.3 | 0.8 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 2.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 6.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 2.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 1.0 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.2 | 4.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 0.6 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 0.6 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.2 | 1.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.2 | 0.7 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 1.5 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 3.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 4.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.9 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 2.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.1 | 1.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.1 | 0.8 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 0.3 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 0.3 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 1.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.7 | GO:1901567 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.1 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 1.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 1.0 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.7 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 3.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0016563 | obsolete transcription activator activity(GO:0016563) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 0.1 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.5 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 3.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0016301 | kinase activity(GO:0016301) |
| 0.0 | 0.8 | GO:0016829 | lyase activity(GO:0016829) |
| 0.0 | 0.4 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 8.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 3.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.8 | GO:0004871 | signal transducer activity(GO:0004871) |
| 0.0 | 3.2 | GO:0030528 | obsolete transcription regulator activity(GO:0030528) |
| 0.0 | 0.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.8 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.1 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.3 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.9 | ST_STAT3_PATHWAY | STAT3 Pathway |
| 0.0 | 2.0 | SIG_PIP3_SIGNALING_IN_B_LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 1.1 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.9 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |